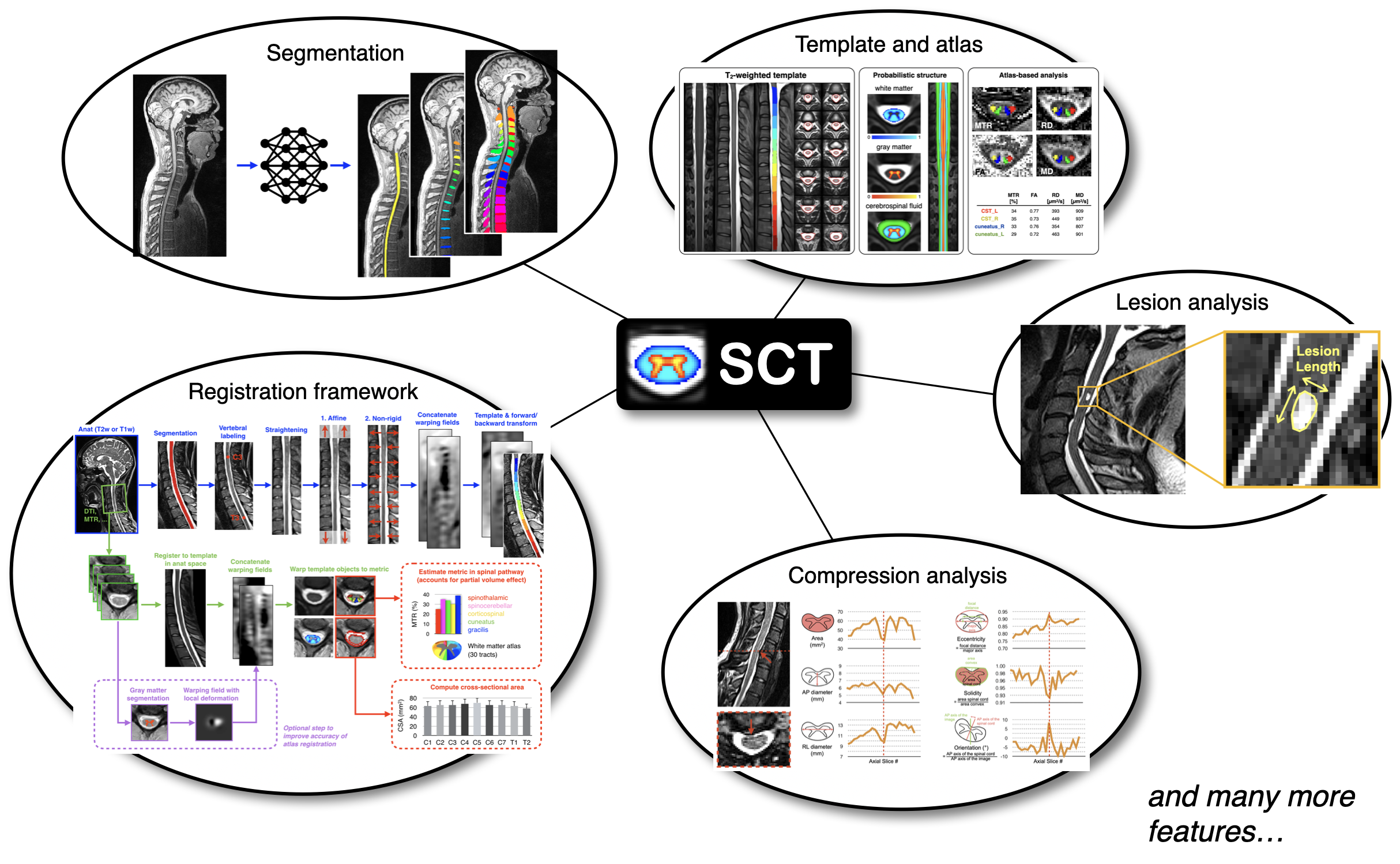
�[1;32mSubject: errsm_23�[0m
�[1;31mContrast: mt�[0m
==============================================================================================
sct_register_multimodal.py
-i /home/django/jcohen2/code/spinalcordtoolbox/data/template/MNI-Poly-AMU_T2.nii.gz
-d /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/mt/mtc1.nii.gz
-s /home/django/jcohen2/code/spinalcordtoolbox/data/template/MNI-Poly-AMU_cord.nii.gz
-t /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/mt/segmentation_binary.nii.gz
-q /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_template2anat.nii.gz
-x 1
-z /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_anat2template.nii.gz
-o template2mtc1.nii.gz
-n 50x20
-r 1
-v 1
==============================================================================================
/home/django/jcohen2/code/spinalcordtoolbox
Check parameters:
.. Source: /home/django/jcohen2/code/spinalcordtoolbox/data/template/MNI-Poly-AMU_T2.nii.gz
.. Destination: /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/mt/mtc1.nii.gz
.. Segmentation source: /home/django/jcohen2/code/spinalcordtoolbox/data/template/MNI-Poly-AMU_cord.nii.gz
.. Segmentation dest: /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/mt/segmentation_binary.nii.gz
.. Init transfo: /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_template2anat.nii.gz
.. Output name: template2mtc1.nii.gz
.. number of iterations: 50x20
.. Verbose: 1
.. Remove temp files: 1
Create local temp files...
>> c3d /home/django/jcohen2/code/spinalcordtoolbox/data/template/MNI-Poly-AMU_T2.nii.gz -o tmp.src.nii
>> c3d /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/mt/mtc1.nii.gz -o tmp.dest.nii
>> c3d /home/django/jcohen2/code/spinalcordtoolbox/data/template/MNI-Poly-AMU_cord.nii.gz -o tmp.src_seg.nii
>> c3d /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/mt/segmentation_binary.nii.gz -o tmp.dest_seg.nii
Apply initial transformation to moving image...
>> WarpImageMultiTransform 3 tmp.src.nii tmp.src_reg.nii -R tmp.dest.nii /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_template2anat.nii.gz --use-BSpline
>> WarpImageMultiTransform 3 tmp.src_seg.nii tmp.src_seg_reg.nii -R tmp.dest_seg.nii /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_template2anat.nii.gz --use-BSpline
Pad source...
>> c3d tmp.src_reg -pad 0x0x3vox 0x0x3vox 0 -o tmp.src_reg_pad.nii
Pad destination...
>> c3d tmp.dest -pad 0x0x3vox 0x0x3vox 0 -o tmp.dest_pad.nii
Pad source segmentation...
>> c3d tmp.src_seg_reg -pad 0x0x3vox 0x0x3vox 0 -o tmp.src_seg_reg_pad.nii
Pad destination segmentation...
>> c3d tmp.dest_seg -pad 0x0x3vox 0x0x3vox 0 -o tmp.dest_seg_pad.nii
Step #1: Estimate transformation using spinal cord segmentations...
>> antsRegistration --dimensionality 3 --transform SyN[0.5,3,0] --metric MI[tmp.dest_seg_pad.nii,tmp.src_seg_reg_pad.nii,1,32] --convergence 50x20 --shrink-factors 4x1 --smoothing-sigmas 1x1mm --Restrict-Deformation 1x1x0 --output [tmp.regSeg,tmp.regSeg.nii]
Using double precision for computations.
number of levels = 2
fixed image: tmp.dest_seg_pad.nii
moving image: tmp.src_seg_reg_pad.nii
Dimension = 3
Number of stages = 1
Use Histogram Matching false
Winsorize Image Intensities false
Lower Quantile = 0
Upper Quantile = 1
Stage 1 State
Metric = Mattes
Fixed Image = Image (0x7fd6e190bb80)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 559
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 396
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
Spacing: [0.84375, 0.84375, 16.5]
Origin: [-81, 117.424, -37.3294]
Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
IndexToPointMatrix:
0.84375 0 0
0 -0.837639 -1.98226
0 -0.101366 16.3805
PointToIndexMatrix:
1.18519 0 0
0 -1.1766 -0.142384
0 -0.00728103 0.0601671
Inverse Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
PixelContainer:
ImportImageContainer (0x7fd6e190baa0)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 393
Debug: Off
Object Name:
Observers:
none
Pointer: 0x108c2d000
Container manages memory: true
Size: 405504
Capacity: 405504
Moving Image = Image (0x7fd6e190b790)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 560
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 557
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
Spacing: [0.84375, 0.84375, 16.5]
Origin: [-81, 117.424, -37.3294]
Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
IndexToPointMatrix:
0.84375 0 0
0 -0.837639 -1.98226
0 -0.101366 16.3805
PointToIndexMatrix:
1.18519 0 0
0 -1.1766 -0.142384
0 -0.00728103 0.0601671
Inverse Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
PixelContainer:
ImportImageContainer (0x7fd6e190fb90)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 554
Debug: Off
Object Name:
Observers:
none
Pointer: 0x10925e000
Container manages memory: true
Size: 405504
Capacity: 405504
Weighting = 1
Sampling Strategy = none
NumberOfBins = 32
Radius = 1
Sampling percentage = 1
Transform = SyN
Gradient Step = 0.5
Update Field Sigma (voxel space) = 3
Total Field Sigma (voxel space) = 0
Update Field Time Sigma = 0
Total Field Time Sigma = 0
Number of Time Indices = 0
Number of Time Point Samples = 0
Registration using 1 total stages.
Stage 0
iterations = 50x20
convergence threshold = 1e-06
convergence window size = 10
number of levels = 2
Shrink factors (level 1 out of 2): [4, 4, 1]
Shrink factors (level 2 out of 2): [1, 1, 1]
smoothing sigmas per level: [1, 1]
using the Mattes MI metric (number of bins = 32, weight = 1)
Using default NONE metricSamplingStrategy
*** Running SyN registration (varianceForUpdateField = 3, varianceForTotalField = 0) ***
Current level = 1 of 2
number of iterations = 50
shrink factors = [4, 4, 1]
smoothing sigmas = 1 mm
required fixed parameters = [48, 48, 11, -79.734375, 116.16756798783919, -37.481424485533395, 3.375, 3.375, 16.499998092651367, 1, 0, 0, 0, -0.9927573344542117, -0.12013689654985334, 0, -0.12013690060663551, 0.9927573349451364]
XXDIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
1DIAGNOSTIC, 1, -1.279534399088e-02, 1.797693134862e+308, 1.2522e-01, 1.2518e-01,
1DIAGNOSTIC, 2, -1.374867870519e-02, 1.797693134862e+308, 1.9391e-01, 6.8693e-02,
1DIAGNOSTIC, 3, -1.495346809446e-02, 1.797693134862e+308, 2.6098e-01, 6.7064e-02,
1DIAGNOSTIC, 4, -1.423497312352e-02, 1.797693134862e+308, 3.2806e-01, 6.7081e-02,
1DIAGNOSTIC, 5, -1.362644975415e-02, 1.797693134862e+308, 3.9576e-01, 6.7702e-02,
1DIAGNOSTIC, 6, -1.273267177939e-02, 1.797693134862e+308, 4.6333e-01, 6.7569e-02,
1DIAGNOSTIC, 7, -1.244140922900e-02, 1.797693134862e+308, 5.3102e-01, 6.7692e-02,
1DIAGNOSTIC, 8, -1.212294379761e-02, 1.797693134862e+308, 5.9814e-01, 6.7124e-02,
1DIAGNOSTIC, 9, -1.192206125885e-02, 1.797693134862e+308, 6.6552e-01, 6.7379e-02,
1DIAGNOSTIC, 10, -1.183634862858e-02, -1.204498820754e-02, 7.3376e-01, 6.8236e-02,
Current level = 2 of 2
number of iterations = 20
shrink factors = [1, 1, 1]
smoothing sigmas = 1.0000e+00 mm
required fixed parameters = [192, 192, 11, -81, 117.42402648925781, -37.329376220703125, 0.84375, 0.84375, 16.499998092651367, 1, 0, 0, 0, -0.9927573344542117, -0.12013689654985334, 0, -0.12013690060663551, 0.9927573349451364]
XXDIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
1DIAGNOSTIC, 1, -1.188294427336e-02, 1.797693134862e+308, 1.7782e+00, 1.0442e+00,
1DIAGNOSTIC, 2, -1.204289498118e-02, 1.797693134862e+308, 2.6767e+00, 8.9846e-01,
1DIAGNOSTIC, 3, -1.212765121139e-02, 1.797693134862e+308, 3.5633e+00, 8.8665e-01,
1DIAGNOSTIC, 4, -1.178380170855e-02, 1.797693134862e+308, 4.4626e+00, 8.9924e-01,
1DIAGNOSTIC, 5, -1.148477022465e-02, 1.797693134862e+308, 5.3465e+00, 8.8390e-01,
1DIAGNOSTIC, 6, -1.122054959059e-02, 1.797693134862e+308, 6.2338e+00, 8.8731e-01,
1DIAGNOSTIC, 7, -1.081726229362e-02, 1.797693134862e+308, 7.0880e+00, 8.5420e-01,
1DIAGNOSTIC, 8, -1.045450851249e-02, 1.797693134862e+308, 7.9820e+00, 8.9406e-01,
1DIAGNOSTIC, 9, -1.017071193630e-02, 1.797693134862e+308, 8.8457e+00, 8.6368e-01,
1DIAGNOSTIC, 10, -9.903069106736e-03, -1.342427600251e-02, 9.7246e+00, 8.7894e-01,
Elapsed time (stage 0): 9.8222e+00
Total elapsed time: 9.8376e+00
Step #2: Improve local deformation using images (start from previous transformation)...
>> antsRegistration --dimensionality 3 --initial-moving-transform tmp.regSeg0Warp.nii.gz --transform SyN[0.1,1,0] --metric MI[tmp.dest_pad.nii,tmp.src_reg_pad.nii,1,32] --convergence 20 --shrink-factors 1 --smoothing-sigmas 0mm --Restrict-Deformation 1x1x0 --output [tmp.reg,tmp.src_reg_pad_reg.nii] --collapse-output-transforms 0 --interpolation BSpline[3]
Using double precision for computations.
=============================================================================
The composite transform is comprised of the following transforms (in order):
1. tmp.regSeg0Warp.nii.gz (type = DisplacementFieldTransform)
=============================================================================
Can't write transform file tmp.reg0DerivedInitialMovingTranslation.mat
Exception Object caught:
itk::ImageFileWriterException (0x7fa83991a998)
Location: "unknown"
File: /home/django/jcohen2/antsbin/ITKv4-install/include/ITK-4.5/itkImageFileWriter.hxx
Line: 151
Description: Could not create IO object for file tmp.reg0DerivedInitialMovingTranslation.mat
Tried to create one of the following:
NiftiImageIO
NrrdImageIO
GiplImageIO
HDF5ImageIO
JPEGImageIO
GDCMImageIO
BMPImageIO
LSMImageIO
PNGImageIO
TIFFImageIO
VTKImageIO
StimulateImageIO
BioRadImageIO
MetaImageIO
MRCImageIO
You probably failed to set a file suffix, or
set the suffix to an unsupported type.
number of levels = 1
fixed image: tmp.dest_pad.nii
moving image: tmp.src_reg_pad.nii
Dimension = 3
Number of stages = 1
Use Histogram Matching false
Winsorize Image Intensities false
Lower Quantile = 0
Upper Quantile = 1
Stage 1 State
Metric = Mattes
Fixed Image = Image (0x7fa83991eec0)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 1488
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1325
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
Spacing: [0.84375, 0.84375, 16.5]
Origin: [-81, 117.424, -37.3294]
Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
IndexToPointMatrix:
0.84375 0 0
0 -0.837639 -1.98226
0 -0.101366 16.3805
PointToIndexMatrix:
1.18519 0 0
0 -1.1766 -0.142384
0 -0.00728103 0.0601671
Inverse Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
PixelContainer:
ImportImageContainer (0x7fa83991e140)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1322
Debug: Off
Object Name:
Observers:
none
Pointer: 0x104077000
Container manages memory: true
Size: 405504
Capacity: 405504
Moving Image = Image (0x7fa83991aa60)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 1489
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1486
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [192, 192, 11]
Spacing: [0.84375, 0.84375, 16.5]
Origin: [-81, 117.424, -37.3294]
Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
IndexToPointMatrix:
0.84375 0 0
0 -0.837639 -1.98226
0 -0.101366 16.3805
PointToIndexMatrix:
1.18519 0 0
0 -1.1766 -0.142384
0 -0.00728103 0.0601671
Inverse Direction:
1 0 0
0 -0.992757 -0.120137
0 -0.120137 0.992757
PixelContainer:
ImportImageContainer (0x7fa83991c7e0)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1483
Debug: Off
Object Name:
Observers:
none
Pointer: 0x1046a7000
Container manages memory: true
Size: 405504
Capacity: 405504
Weighting = 1
Sampling Strategy = none
NumberOfBins = 32
Radius = 1
Sampling percentage = 1
Transform = SyN
Gradient Step = 0.1
Update Field Sigma (voxel space) = 1
Total Field Sigma (voxel space) = 0
Update Field Time Sigma = 0
Total Field Time Sigma = 0
Number of Time Indices = 0
Number of Time Point Samples = 0
Registration using 1 total stages.
Stage 0
iterations = 20
convergence threshold = 1e-06
convergence window size = 10
number of levels = 1
Shrink factors (level 1 out of 1): [1, 1, 1]
smoothing sigmas per level: [0]
using the Mattes MI metric (number of bins = 32, weight = 1)
Using default NONE metricSamplingStrategy
*** Running SyN registration (varianceForUpdateField = 1, varianceForTotalField = 0) ***
Current level = 1 of 1
number of iterations = 20
shrink factors = [1, 1, 1]
smoothing sigmas = 0 mm
required fixed parameters = [192, 192, 11, -81, 117.42402648925781, -37.329376220703125, 0.84375, 0.84375, 16.499998092651367, 1, 0, 0, 0, -0.9927573344542117, -0.12013688942932219, 0, -0.12013690060663551, 0.9927573358068157]
XXDIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
1DIAGNOSTIC, 1, -7.405185984578e-02, 1.797693134862e+308, 8.4241e-01, 8.4238e-01,
1DIAGNOSTIC, 2, -7.416685794359e-02, 1.797693134862e+308, 1.6306e+00, 7.8823e-01,
1DIAGNOSTIC, 3, -7.429667691027e-02, 1.797693134862e+308, 2.4292e+00, 7.9856e-01,
1DIAGNOSTIC, 4, -7.443575182016e-02, 1.797693134862e+308, 3.2509e+00, 8.2173e-01,
1DIAGNOSTIC, 5, -7.453851632370e-02, 1.797693134862e+308, 4.0509e+00, 7.9996e-01,
1DIAGNOSTIC, 6, -7.462891831747e-02, 1.797693134862e+308, 4.8645e+00, 8.1358e-01,
1DIAGNOSTIC, 7, -7.467508336003e-02, 1.797693134862e+308, 5.6534e+00, 7.8892e-01,
1DIAGNOSTIC, 8, -7.470231353181e-02, 1.797693134862e+308, 6.4346e+00, 7.8122e-01,
1DIAGNOSTIC, 9, -7.477296301922e-02, 1.797693134862e+308, 7.2420e+00, 8.0737e-01,
1DIAGNOSTIC, 10, -7.481694391767e-02, 6.511445570990e-04, 8.0645e+00, 8.2253e-01,
1DIAGNOSTIC, 11, -7.482867220136e-02, 4.985201012338e-04, 8.8669e+00, 8.0243e-01,
1DIAGNOSTIC, 12, -7.490023421227e-02, 3.859223037805e-04, 9.6761e+00, 8.0917e-01,
1DIAGNOSTIC, 13, -7.493715113159e-02, 3.037841859762e-04, 1.0493e+01, 8.1642e-01,
1DIAGNOSTIC, 14, -7.500837784343e-02, 2.623704060583e-04, 1.1312e+01, 8.1950e-01,
1DIAGNOSTIC, 15, -7.499624945127e-02, 2.248124841403e-04, 1.2135e+01, 8.2301e-01,
1DIAGNOSTIC, 16, -7.506446438221e-02, 2.094209181930e-04, 1.2946e+01, 8.1082e-01,
1DIAGNOSTIC, 17, -7.504515794096e-02, 1.822666293318e-04, 1.3760e+01, 8.1441e-01,
1DIAGNOSTIC, 18, -7.510618672287e-02, 1.597023041366e-04, 1.4573e+01, 8.1294e-01,
1DIAGNOSTIC, 19, -7.513799412739e-02, 1.448281665660e-04, 1.5380e+01, 8.0657e-01,
1DIAGNOSTIC, 20, -7.515362088850e-02, 1.294023752298e-04, 1.6188e+01, 8.0837e-01,
Elapsed time (stage 0): 1.6289e+01
Total elapsed time: 1.6304e+01
Concatenate transformations...
>> ComposeMultiTransform 3 tmp.warp_src2dest.nii.gz -R tmp.dest.nii tmp.reg1Warp.nii.gz tmp.regSeg0Warp.nii.gz /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_template2anat.nii.gz
>> ComposeMultiTransform 3 tmp.warp_dest2src.nii.gz -R tmp.src.nii /home/django/jcohen2/code/spinalcordtoolbox/testing/data/errsm_23/template/warp_anat2template.nii.gz tmp.regSeg0InverseWarp.nii.gz tmp.reg1InverseWarp.nii.gz
Apply transfo source --> dest...
>> WarpImageMultiTransform 3 tmp.src.nii tmp.src_reg.nii -R tmp.dest.nii tmp.warp_src2dest.nii.gz --use-BSpline
Apply transfo dest --> source...
>> WarpImageMultiTransform 3 tmp.dest.nii tmp.dest_reg.nii -R tmp.src.nii tmp.warp_dest2src.nii.gz --use-BSpline
ERROR!!!
FIELD: tmp.warp_dest2src.nii.gz
moving_image_filename: tmp.dest.nii components 1
output_image_filename: tmp.dest_reg.nii
reference_image_filename: tmp.src.nii
[0/1]: FIELD: tmp.warp_dest2src.nii.gz
Not currently supported because of a lack of vector support
Exception caught during WarpImageMultiTransform.
itk::ImageFileReaderException (0x7f8b04912688)
Location: "unknown"
File: /home/django/jcohen2/antsbin/ITKv4-install/include/ITK-4.5/itkImageFileReader.hxx
Line: 143
Description: Could not create IO object for file tmp.warp_dest2src.nii.gz
The file doesn't exist.
Filename = tmp.warp_dest2src.nii.gz
Exit program.