This repo contains both a specification for a JSON/YAML format for ontology exchange, plus a reference java object model and OWL converter.
The core is a simple graph model allowing the expression of ontology
relationships like forelimb SubClassOf limb:
"nodes" : [
{
"id" : "UBERON:0002102",
"lbl" : "forelimb"
}, {
"id" : "UBERON:0002101",
"lbl" : "limb"
}
],
"edges" : [
{
"subj" : "UBERON:0002102",
"pred" : "is_a",
"obj" : "UBERON:0002101"
}
]
Additional optional fields allow increased expressivity, without adding complexity to the core.
For more examples, see examples/ in this repo - or for real-world examples, this drive. Soon we hope to have this incorporated into release tools and visible at standard PURLs.
For the JSON Schema, see the schema/ folder
If you are familiar with OWL, skip straight to the OWL mapping specification
Currently if a developer needs to add ontologies into a software framework or tool, there are two options for formats: obo-format and OWL (technically obo is an OWL syntax, but for pragmatic purposes we can separate these two).
This presents a number of problems: obo is simple, but employs its own syntax, resulting in a proliferation of ad-hoc parsers that are generally incomplete. It is also less expressive than OWL (but expressive enough for the majority of bioinformatics tasks). OWL is a W3 standard, but can be difficult to work with. Typically OWL is layered on RDF, but RDF level libraries can be too low-level to work with (additionally: rdflib for Python is very slow). For JVM languages, the OWLAPI can be used, but this can be abstruse for many routine tasks, leading to variety of simplifying facades each with their own assumptions (e.g. BRAIN).
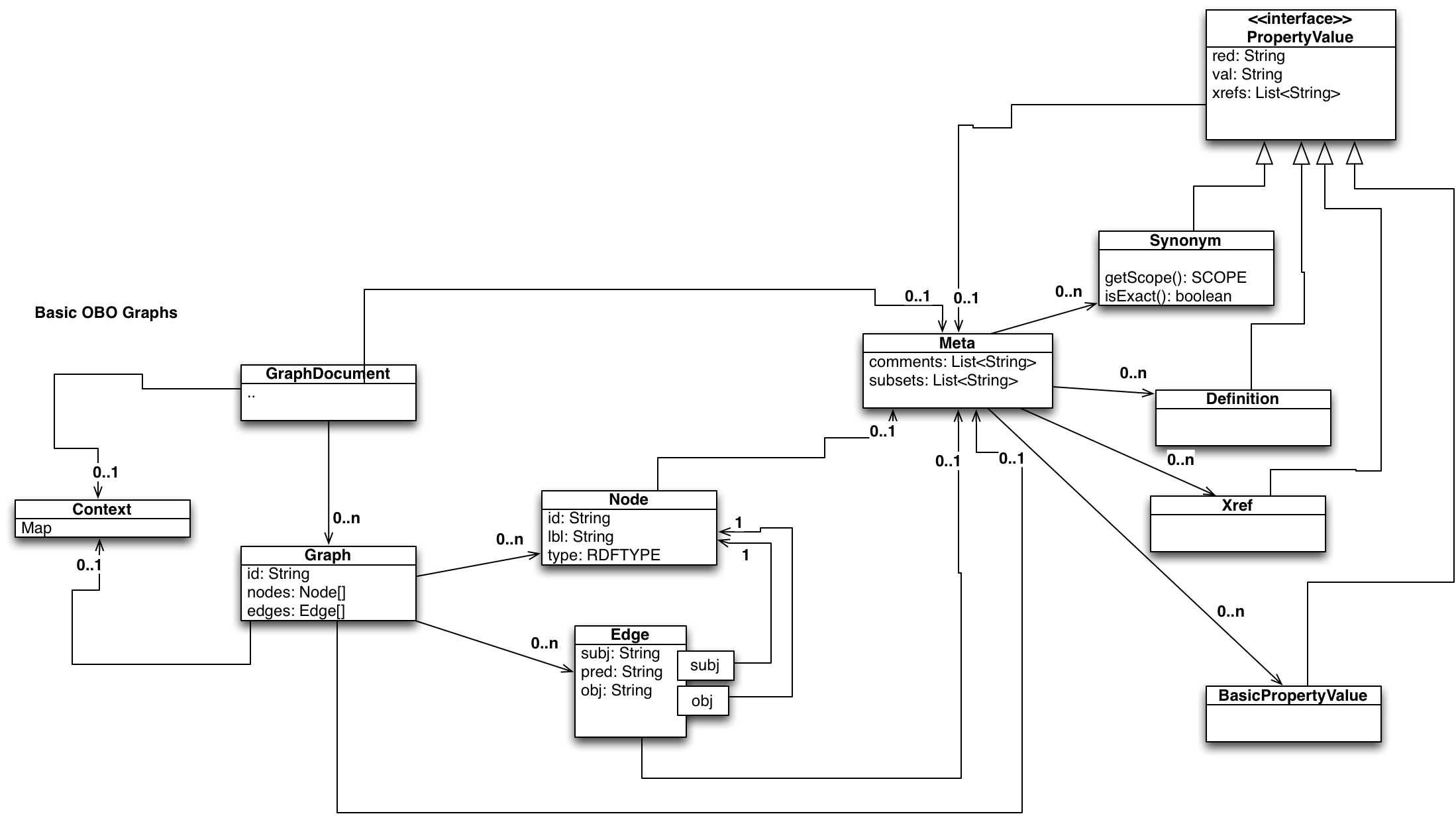
OBO Graphs (OGs) are a graph-oriented way of representing ontologies or portions of ontologies in a developer-friendly JSON (or YAML) format. A typical consumer may be a Python developer using ontologies to enhance an analysis tool, database search/infrastructure etc.
The model can be understood as two levels: A basic level, that is intended to satisfy 99% of bioinformatics use cases, and is essentially a cytoscape-like nodes and edges model. On top of this is an expressive level that allows the representation of more esoteric OWL axioms.
The core model is a property-labeled graph, comparable to the data model underlying graph databases such as Neo4j. The format is the same as BBOP-Graphs.
The basic form is:
"graphs": [
{
"nodes" : [...],
"edges" : [
],
},
...
]
Here is an example of a subgraph of Uberon consisting of four nodes, two part-of and two is_a edges:
{
"nodes" : [
{
"id" : "UBERON:0002470",
"lbl" : "autopod region"
}, {
"id" : "UBERON:0002102",
"lbl" : "forelimb"
}, {
"id" : "UBERON:0002101",
"lbl" : "limb"
}, {
"id" : "UBERON:0002398",
"lbl" : "manus"
}
],
"edges" : [
{
"subj" : "UBERON:0002102",
"pred" : "is_a",
"obj" : "UBERON:0002101"
}, {
"subj" : "UBERON:0002398",
"pred" : "part_of",
"obj" : "UBERON:0002102"
}, {
"subj" : "UBERON:0002398",
"pred" : "is_a",
"obj" : "UBERON:0002470"
}, {
"subj" : "UBERON:0002470",
"pred" : "part_of",
"obj" : "UBERON:0002101"
}
]
}
The short forms in the above (e.g. UBERON:0002470 and part_of) are
mapped to unambiguous PURLs using a JSON-LD context (see below).
Edges can also be decorated with Meta objects (corresponding to
reification in RDF/OWL, or edge properties in graph databases).
Formally, the set of edges correspond to OWL SubClassOf axioms of two forms:
C SubClassOf D(akais_ain obo-format)C SubClassOf P some D(akarelationshipin obo-format)
For a full description, see the JSON Schema below
Nodes collect all OWL annotations about an entity.
Typically nodes will be OWL classes, but they can also be OWL individuals, or OWL properties (in which case edges can also correspond to SubPropertyOf axioms)
Nodes, edges and graphs can have optional meta objects for
additional metadata (or annotations in OWL speak).
Here is an example of a meta object for a GO class (show in YAML, for compactness):
- id: "http://purl.obolibrary.org/obo/GO_0044464"
meta:
definition:
val: "Any constituent part of a cell, the basic structural and functional\
\ unit of all organisms."
xrefs:
- "GOC:jl"
subsets:
- "http://purl.obolibrary.org/obo/go/subsets/nucleus#goantislim_grouping"
- "http://purl.obolibrary.org/obo/go/subsets/nucleus#gosubset_prok"
- "http://purl.obolibrary.org/obo/go/subsets/nucleus#goslim_pir"
- "http://purl.obolibrary.org/obo/go/subsets/nucleus#gocheck_do_not_annotate"
xrefs:
- val: "NIF_Subcellular:sao628508602"
synonyms:
- pred: "hasExactSynonym"
val: "cellular subcomponent"
xrefs:
- "NIF_Subcellular:sao628508602"
- pred: "hasRelatedSynonym"
val: "protoplast"
xrefs:
- "GOC:mah"
type: "CLASS"
lbl: "cell part"
These provide ways of expressing logical axioms not covered in the subset above.
Currently the spec does not provide a complete translation of all OWL axioms. This will be driven by comments on the spec.
Currently two axiom patterns are defined:
- equivalenceSet
- logicalDefinitionAxiom
Note that these do not necessarily correspond 1:1 to OWL axiom types. The two above are different forms of equivalent classes axiom, the former suited to cases where we have multiple ontologies with the same concept represented using a different URI in each (for example, a DOID:nnn URI and a Orphanet:nnn URI with a direct equivalence axiom between them).
The latter is for so called 'cross-product' or 'genus-differentia' definitions found in most well-behaved bio-ontologies.
See README-owlmapping.md for mor details
See bbop-graph
- Top-level object in a bbop-graph is a
graphobject; in obographs aGraphDocumentis a holder for multiple graphs metaobjects are underspecified in bbop-graphs
See Neo4jMapping
The mapping is similar, particularly with respect to how SubClassOf axioms map to edges. However, for SciGraph, more advanced axioms such as EquivalenceAxioms are mapped to graph edges. In obographs, anything outside the BOG pattern is mapped to a custom object.
Note also that SciGraph returns bbop-graph objects by default from graph query operations.
mvn install
./bin/ogger src/test/resources/basic.obo
Note that the conversion will be rolled into tools like ROBOT obviating the need for this. We can also make it such that the JSON is available from a standard PURL, e.g.
- http://purl.obolibrary.org/obo/envo.obo
- http://purl.obolibrary.org/obo/envo.owl
- http://purl.obolibrary.org/obo/envo.json NEW
The library is split into two modules - obographs-core which contains the model and code for reading and writing JSON
and YAML graphs. The obographs-owlapi requires obographs-core and includes the owlapi and code for converting OWL to
obographs.
<dependency>
<groupId>org.geneontology.obographs</groupId>
<artifactId>obographs-core</artifactId>
<version>${project.version}</version>
</dependency>compile 'org.geneontology.obographs:obographs-core:${project.version}'When developing against an unreleased snapshot version of the API, you can use Maven to install it in your local m2 repository:
mvn clean install
If you find that your IDE cannot load any of the concrete classes e.g. Graph or GraphDocument you should check that
your IDE has Annotation Processing enabled. Obographs uses the immutables library which
requires annotation processing in the IDE. See https://immutables.github.io/apt.html for how to enable this in your IDE.
You might need to restart your IDE or re-import the maven projects for this to work fully. It is not required for projects
using obographs as a pre-built library.
mvn clean deploy -P release
See bbop-graph