This is a Python re-implementation of the spectral clustering algorithms presented in these papers:
| Algorithm | Paper |
|---|---|
| Refined Laplacian matrix | Speaker Diarization with LSTM |
| Constrained spectral clustering | Turn-to-Diarize: Online Speaker Diarization Constrained by Transformer Transducer Speaker Turn Detection |
| Multi-stage clustering | Highly Efficient Real-Time Streaming and Fully On-Device Speaker Diarization with Multi-Stage Clustering |
We recently added new functionalities to this library to include algorithms in a new paper. We updated the APIs as well.
If you depend on our old API, please use an older version of this library:
pip3 install spectralcluster==0.1.0
This is not a Google product.
This is not the original C++ implementation used by the papers.
Please consider this repo as a "demonstration" of the algorithms, instead of a "reproduction" of what we use at Google. Some features might be missing or incomplete.
Install the package by:
pip3 install spectralclusteror
python3 -m pip install spectralclusterSimply use the predict() method of class SpectralClusterer to perform
spectral clustering. The example below should be closest to the original C++
implemention used by our
ICASSP 2018 paper.
from spectralcluster import configs
labels = configs.icassp2018_clusterer.predict(X)The input X is a numpy array of shape (n_samples, n_features),
and the returned labels is a numpy array of shape (n_samples,).
You can also create your own clusterer like this:
from spectralcluster import SpectralClusterer
clusterer = SpectralClusterer(
min_clusters=2,
max_clusters=7,
autotune=None,
laplacian_type=None,
refinement_options=None,
custom_dist="cosine")
labels = clusterer.predict(X)
For the complete list of parameters of SpectralClusterer, see
spectralcluster/spectral_clusterer.py.
In our ICASSP 2018 paper, we apply a sequence of refinment operations on the affinity matrix, which is critical to the performance on the speaker diarization results.
You can specify your refinement operations like this:
from spectralcluster import RefinementOptions
from spectralcluster import ThresholdType
from spectralcluster import ICASSP2018_REFINEMENT_SEQUENCE
refinement_options = RefinementOptions(
gaussian_blur_sigma=1,
p_percentile=0.95,
thresholding_soft_multiplier=0.01,
thresholding_type=ThresholdType.RowMax,
refinement_sequence=ICASSP2018_REFINEMENT_SEQUENCE)
Then you can pass the refinement_options as an argument when initializing your
SpectralClusterer object.
For the complete list of RefinementOptions, see
spectralcluster/refinement.py.
In our ICASSP 2018 paper,
we apply a refinement operation CropDiagonal on the affinity matrix, which replaces each diagonal element of the affinity matrix by the max non-diagonal value of the row. After this operation, the matrix has similar properties to a standard Laplacian matrix, and it is also less sensitive (thus more robust) to the Gaussian blur operation than a standard Laplacian matrix.
In the new version of this library, we support different types of Laplacian matrix now, including:
- None Laplacian (affinity matrix):
W - Unnormalized Laplacian:
L = D - W - Graph cut Laplacian:
L' = D^{-1/2} * L * D^{-1/2} - Random walk Laplacian:
L' = D^{-1} * L
You can specify the Laplacian matrix type with the laplacian_type argument of the SpectralClusterer class.
Note: Refinement operations are applied to the affinity matrix before computing the Laplacian matrix.
In our ICASSP 2018 paper, the K-Means is based on Cosine distance.
You can set custom_dist="cosine" when initializing your SpectralClusterer object.
You can also use other distances supported by scipy.spatial.distance, such as "euclidean" or "mahalanobis".
In our ICASSP 2018 paper,
the affinity between two embeddings is defined as (cos(x,y)+1)/2.
You can also use other affinity functions by setting affinity_function when initializing your SpectralClusterer object.
We also support auto-tuning the p_percentile parameter of the RowWiseThreshold refinement operation, which was original proposed in this paper.
You can enable this by passing in an AutoTune object to the autotune argument when initializing your SpectralClusterer object.
Example:
from spectralcluster import AutoTune, AutoTuneProxy
autotune = AutoTune(
p_percentile_min=0.60,
p_percentile_max=0.95,
init_search_step=0.01,
search_level=3,
proxy=AutoTuneProxy.PercentileSqrtOverNME)For the complete list of parameters of AutoTune, see
spectralcluster/autotune.py.
Spectral clustering exploits the global structure of the data. But there are cases where spectral clustering does not work as well as some other simpler clustering methods, such as when the number of embeddings is too small.
When initializing the SpectralClusterer object, you can pass in a FallbackOptions object to the fallback_options argument, to use a fallback clusterer under certain conditions.
Also, spectral clustering and eigen-gap may not work well at making single-vs-multi cluster decisions. When min_clusters=1, we can also specify FallbackOptions.single_cluster_condition and FallbackOptions.single_cluster_affinity_threshold to help determine single cluster cases by thresdholding the affinity matrix.
For the complete list of parameters of FallbackOptions, see spectralcluster/fallback_clusterer.py.
Spectral clustering can become slow when the number of input embeddings is large. This is due to the high costs of steps such as computing the Laplacian matrix, and eigen decomposition of the Laplacian matrix. One trick to speed up the spectral clustering when the input size is large is to use hierarchical clustering as a pre-clustering step.
To use this feature, you can specify the max_spectral_size argument when constructing the SpectralClusterer object. For example, if you set max_spectral_size=200, then the Laplacian matrix can be at most 200 * 200.
But please note that setting max_spectral_size may cause degradations of the final clustering quality. So please use this feature wisely.
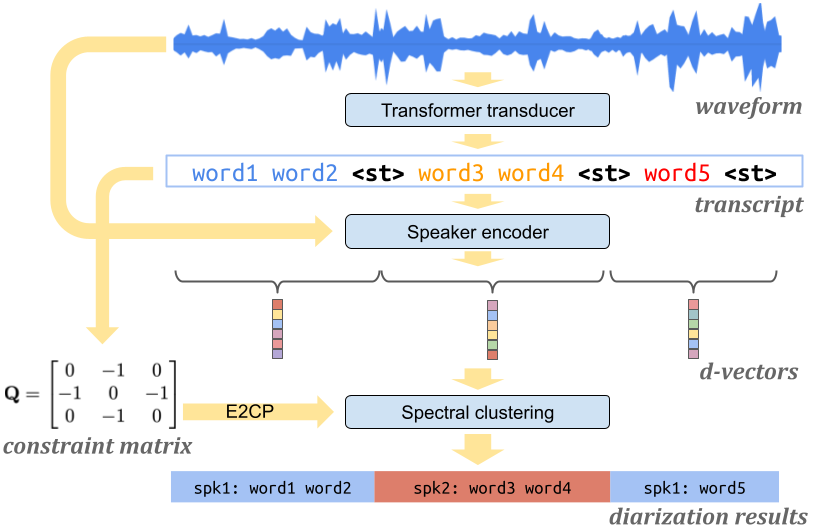
In the Turn-to-Diarize paper, the spectral clustering is constrained by speaker turns. We implemented two constrained spectral clustering methods:
If you pass in a ConstraintOptions object when initializing your SpectralClusterer object, you can call the predict function with a constraint_matrix.
Example usage:
from spectralcluster import constraint
ConstraintName = constraint.ConstraintName
constraint_options = constraint.ConstraintOptions(
constraint_name=ConstraintName.ConstraintPropagation,
apply_before_refinement=True,
constraint_propagation_alpha=0.6)
clusterer = spectral_clusterer.SpectralClusterer(
max_clusters=2,
refinement_options=refinement_options,
constraint_options=constraint_options,
laplacian_type=LaplacianType.GraphCut,
row_wise_renorm=True)
labels = clusterer.predict(matrix, constraint_matrix)The constraint matrix can be constructed from a speaker_turn_scores list:
from spectralcluster import constraint
constraint_matrix = constraint.ConstraintMatrix(
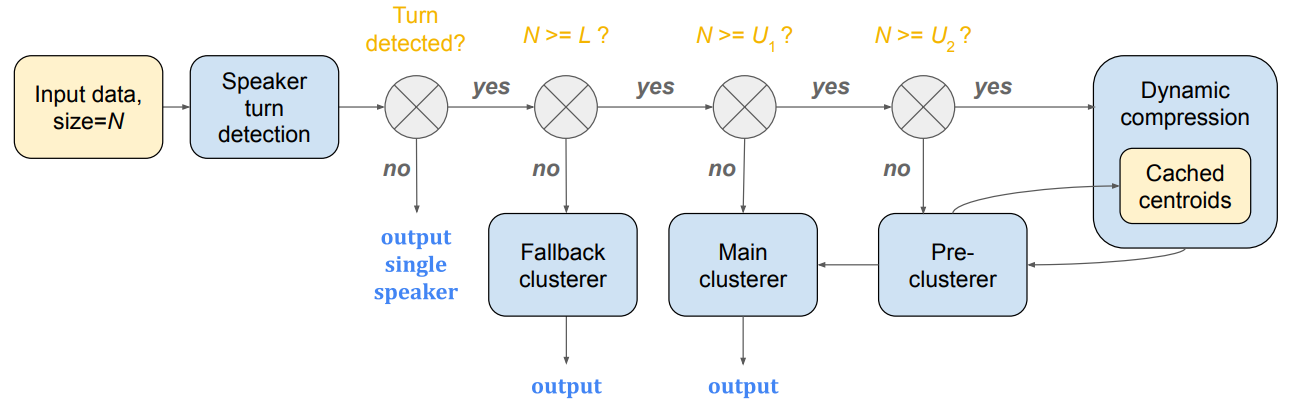
speaker_turn_scores, threshold=1).compute_diagonals()In the multi-stage clustering paper,
we introduced a highly efficient streaming clustering approach. This is
implemented as the MultiStageClusterer class in
spectralcluster/multi_stage_clusterer.py.
Note: We did NOT implement speaker turn detection in this open source library. We only implemented fallback, main, pre-clusterer and dynamic compression here.
The MultiStageClusterer class has a method named streaming_predict.
In streaming clustering, every time we feed a single new embedding to the
streaming_predict function, it will return the sequence of cluster labels
for all inputs, including corrections for the predictions on previous
embeddings.
Example usage:
from spectralcluster import Deflicker
from spectralcluster import MultiStageClusterer
from spectralcluster import SpectralClusterer
main_clusterer = SpectralClusterer()
multi_stage = MultiStageClusterer(
main_clusterer=main_clusterer,
fallback_threshold=0.5,
L=50,
U1=200,
U2=400,
deflicker=Deflicker.Hungarian)
for embedding in embeddings:
labels = multi_stage.streaming_predict(embedding)Our papers are cited as:
@inproceedings{wang2018speaker,
title={{Speaker Diarization with LSTM}},
author={Wang, Quan and Downey, Carlton and Wan, Li and Mansfield, Philip Andrew and Moreno, Ignacio Lopz},
booktitle={2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)},
pages={5239--5243},
year={2018},
organization={IEEE}
}
@inproceedings{xia2022turn,
title={{Turn-to-Diarize: Online Speaker Diarization Constrained by Transformer Transducer Speaker Turn Detection}},
author={Wei Xia and Han Lu and Quan Wang and Anshuman Tripathi and Yiling Huang and Ignacio Lopez Moreno and Hasim Sak},
booktitle={2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)},
pages={8077--8081},
year={2022},
organization={IEEE}
}
@article{wang2022highly,
title={Highly Efficient Real-Time Streaming and Fully On-Device Speaker Diarization with Multi-Stage Clustering},
author={Quan Wang and Yiling Huang and Han Lu and Guanlong Zhao and Ignacio Lopez Moreno},
journal={arXiv:2210.13690},
year={2022}
}
We also have fully supervised speaker diarization systems, powered by uis-rnn. Check this Google AI Blog.
Also check out our recent work on DiarizationLM.
To learn more about speaker diarization, you can check out:
- A curated list of resources: awesome-diarization
- An online course on Udemy: A Tutorial on Speaker Diarization