Chisio BioPAX Editor, or simply ChiBE, is a free editing and visualization tool for pathway models represented by the BioPAX format, using SBGN Process Description Language, based on Chisio.
The tool features user-friendly display, viewing, and editing of pathway models. Pathway views are rendered in a feature-rich format, and may be laid out and edited with state-of-the-art layout and visualization technologies. In addition, facilities for querying a set of integrated pathways in Pathway Commons are provided. Furthermore, visualization of experimental data, including cancer genomics data of the cBioPortal, overlaid on pathway views is supported in ChiBE. These capabilities are organized around a genomics-oriented workflow and offer a unique comprehensive pathway analysis solution for genomics researchers.
ChiBE is a Java application running on various platforms including Windows XP/Vista/7, Linux, and Mac OS X. It was built using Chisio 2.0 and Eclipse GEF 3.1 for graph visualization, Paxtools for accessing and manipulating BioPAX files, and PATIKAmad for experiment data analysis.
For a quick guide on how to explore pathways in Pathway Commons, please see this wiki page.
ChiBE is distributed under Eclipse Public License.
Below are latest builds of ChiBE.
Below are the latest releases of ChiBE 2.2 for different platforms.
Please contact us if you need a build for another platform.
Please see the setup guide for running ChiBE from sources, and quick start guide for fast examples.
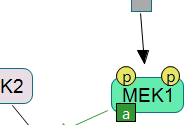
Following are sample screenshots showing tool highlights.
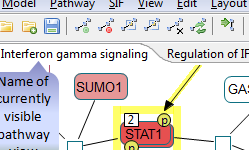 |
 |
Click on the figure to see the entire screenshot.
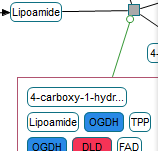
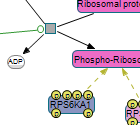
Following are images of pathway models produced by ChiBE taken from some prominent databases.
|
 |
 |
Click on the figure to see the entire screenshot.
(left) ERK/MAPK targets from Reactome (middle) Class IB PI3K non-lipid kinase events from NCI PID (right) Hedgehog signaling pathway from The Cancer Cell Map
- Made architectural improvements in the core.
- Added a Data Legend for the color-coding of the currently loaded experiment data.
- Implemented an enriched-reactions query to rank reactions with a statistical measure.
- Upgraded Pathway Commons interface to the latest version.
- Added support for showing/hiding compartments.
- Revised binary interaction types for SIF views.
- BioPAX level 3 Support: Support for BioPAX level 3 models as well as level 2 models is now provided seamlessly in ChiBE.
- SBGN Process Description Language Compliance: Loaded pathway models are now rendered using a language mostly compliant with SBGN's Process Description Language.
- Remote Querying: Now ChiBE can send graph queries to Pathway Commons database.
- Local Querying: Loaded pathway models can now be queried to search for local relationships.
- Fetching From GEO: ChiBE now offers an alternative way for visualizing expression data on pathways, if an expression data stored in NCBI GEO is to be visualized on a pathway from Pathway Commons. With providing the GEO accession number of series file only, user will be able to visualize the particular expression set in ChiBE.
- Fetching From cBio Portal: cBio Cancer Genomics Portal is a cancer genomics analysis portal, which offers the chance to explore cancer genomes with respect to mutations, copy number alterations, mRNA expression changes, DNA methylation values, and protein levels. ChiBE enables users analysis of such data at pathway level. It collects data from cBio Portal and integrates with the model to display the alterations associated with each entity.
- Upgraded Chisio, ChiBE rendering and layout engine, to version 2.0.
- Support for Mac OS X
- Improved persistence of pathway views as image
- Several minor bug fixes
- Ozgun Babur, Emek Demir, B.Arman Aksoy, Nikolaus Schultz, Chris Sander, cbio at MSKCC
- Ugur Dogrusoz, Merve Cakir, i-Vis at Bilkent University
ChiBE's requirements were determined by OB, UD, ED, NS, and CS. The software design and development were mainly performed by OB, with help from MC, AA, and UD.
We would like to thank IntelliJ for providing a free open source license for IDEA.
Babur, Ö., Dogrusoz, U., Çakir, M., Aksoy, B. A., Schultz, N., Sander, C., & Demir, E. (2014). Integrating biological pathways and genomic profiles with ChiBE 2. BMC genomics, 15(1), 642.
Babur, O., Dogrusoz, U., Demir, E., & Sander, C. (2010). ChiBE: interactive visualization and manipulation of BioPAX pathway models. Bioinformatics, 26(3), 429-431.

