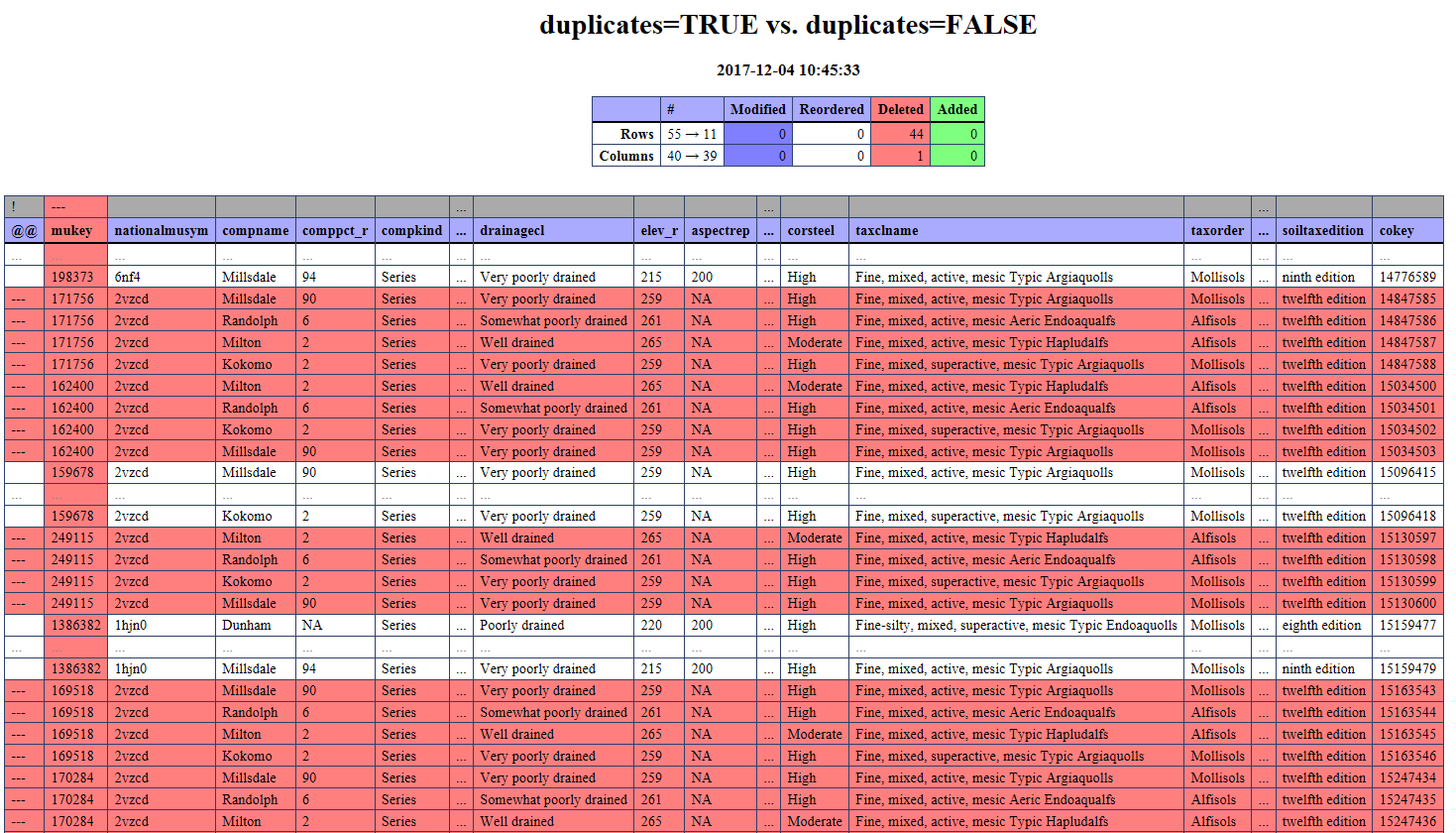
Trying to get my head around this one.
An example from Stephen:
library(soilDB)
library(daff)
# test both cases, thanks Stephen for the example
x <- get_component_from_SDA(WHERE = "muname = 'Millsdale silty clay loam, 0 to 2 percent slopes'", duplicates = TRUE)
y <- get_component_from_SDA(WHERE = "muname = 'Millsdale silty clay loam, 0 to 2 percent slopes'", duplicates = FALSE)
# get differences
d <- diff_data(x, y)
render_diff(d, title='duplicates=TRUE vs. duplicates=FALSE')
The two calls to get_component_from_SDA() generate the following SQL:
-- duplicates == TRUE
SELECT mukey, mu.nationalmusym, compname, comppct_r, compkind, majcompflag, localphase, slope_r, tfact, wei, weg, drainagecl, elev_r, aspectrep, map_r, airtempa_r, reannualprecip_r, ffd_r, nirrcapcl, nirrcapscl, irrcapcl, irrcapscl, frostact, hydgrp, corcon, corsteel, taxclname, taxorder, taxsuborder, taxgrtgroup, taxsubgrp, taxpartsize, taxpartsizemod, taxceactcl, taxreaction, taxtempcl, taxmoistscl, taxtempregime, soiltaxedition, cokey
FROM legend l
INNER JOIN
mapunit mu ON mu.lkey = l.lkey
INNER JOIN (
SELECT compname, comppct_r, compkind, majcompflag, localphase, slope_r, tfact, wei, weg, drainagecl, elev_r, aspectrep, map_r, airtempa_r, reannualprecip_r, ffd_r, nirrcapcl, nirrcapscl, irrcapcl, irrcapscl, frostact, hydgrp, corcon, corsteel, taxclname, taxorder, taxsuborder, taxgrtgroup, taxsubgrp, taxpartsize, taxpartsizemod, taxceactcl, taxreaction, taxtempcl, taxmoistscl, taxtempregime, soiltaxedition, cokey , mukey AS mukey2 FROM component
) AS c ON c.mukey2 = mu.mukey
WHERE muname = 'Millsdale silty clay loam, 0 to 2 percent slopes'
ORDER BY cokey, compname, comppct_r DESC;
-- duplicates == FALSE
SELECT DISTINCT mu.nationalmusym, compname, comppct_r, compkind, majcompflag, localphase, slope_r, tfact, wei, weg, drainagecl, elev_r, aspectrep, map_r, airtempa_r,
reannualprecip_r, ffd_r, nirrcapcl, nirrcapscl, irrcapcl, irrcapscl, frostact, hydgrp, corcon, corsteel, taxclname, taxorder, taxsuborder, taxgrtgroup, taxsubgrp,
taxpartsize, taxpartsizemod, taxceactcl, taxreaction, taxtempcl, taxmoistscl, taxtempregime, soiltaxedition, cokey
FROM legend l
INNER JOIN mapunit mu ON mu.lkey = l.lkey
INNER JOIN (
SELECT MIN(nationalmusym) nationalmusym2, MIN(mukey) AS mukey2
FROM mapunit
GROUP BY nationalmusym
) AS mu2 ON mu2.nationalmusym2 = mu.nationalmusym
INNER JOIN (
SELECT compname, comppct_r, compkind, majcompflag, localphase, slope_r, tfact, wei, weg, drainagecl, elev_r, aspectrep, map_r, airtempa_r, reannualprecip_r,
ffd_r, nirrcapcl, nirrcapscl, irrcapcl, irrcapscl, frostact, hydgrp, corcon, corsteel, taxclname, taxorder, taxsuborder, taxgrtgroup, taxsubgrp, taxpartsize,
taxpartsizemod, taxceactcl, taxreaction, taxtempcl, taxmoistscl, taxtempregime, soiltaxedition, cokey , mukey AS mukey2
FROM component
) AS c ON c.mukey2 = mu2.mukey2
WHERE muname = 'Millsdale silty clay loam, 0 to 2 percent slopes'
ORDER BY cokey, compname, comppct_r DESC
The differences in results, truncated for clarity: