Smoothed particle hydrodynamics analysis and visualization with Python.
Plonk is a Python tool for analysis and visualization of smoothed particle hydrodynamics data with a focus on astrophysical fluid dynamics.
With Plonk we aim to integrate the high quality SPH visualisation of Splash into the modern Python astronomer workflow, and provide a framework for analysis of smoothed particle hydrodynamics simulation data.
Plonk supports the following SPH file formats:
Note: you can convert Phantom non-HDF5 snapshots to HDF5. See the Phantom docs.
To read in a simulation with snapshot files like disc_00000.h5, and global quantity time series files like disc01.ev, in the current directory, and see what snapshots there are:
>>> import plonk
>>> simulation = plonk.load_sim(prefix='disc')
>>> simulation.snaps
[<plonk.Snap "disc_00000.h5">,
...
<plonk.Snap "disc_00030.h5">]You can load individual snapshots and access the particle arrays:
>>> snap = plonk.load_snap(filename='disc_00030.h5')
>>> snap['position']
array([[-3.69505001e+12, 7.42032967e+12, -7.45096980e+11],
...,
[ 1.21421196e+12, 2.08618956e+13, 1.12998892e+12]]) <Unit('meter')>The Snap objects contain the particle arrays, lazily loaded from the HDF5 file, as well as simulation metadata properties stored as a dictionary.
To visualize the column density on a snapshot:
>>> snap.image(quantity='density')
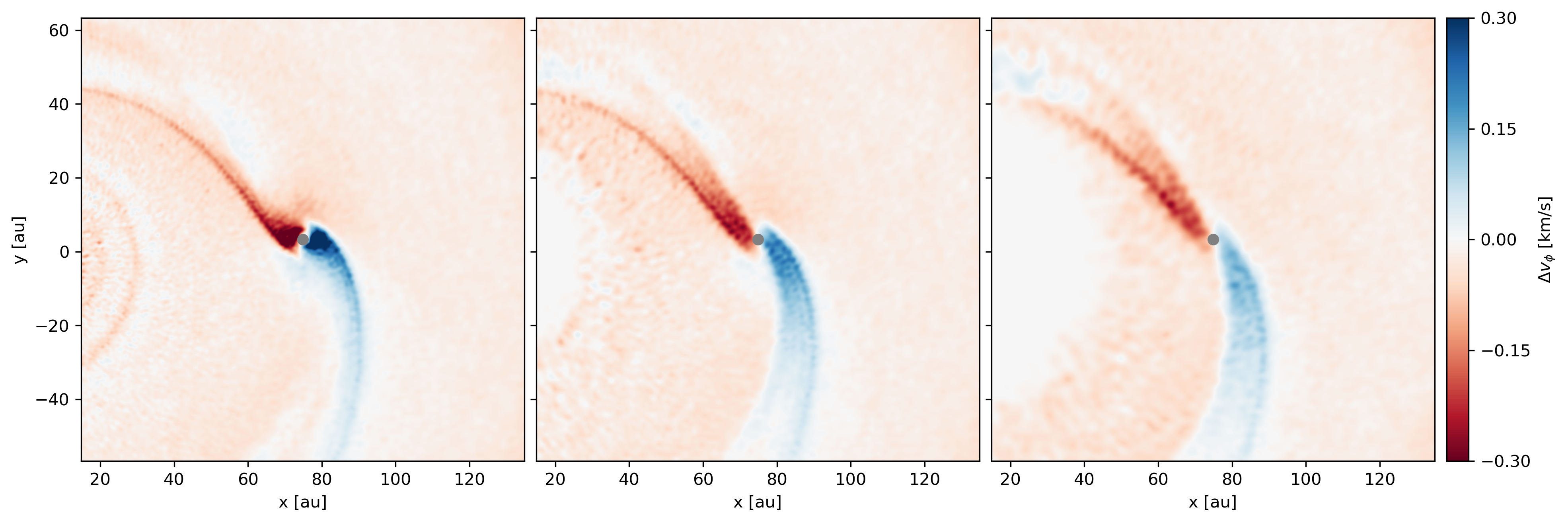
<AxesSubplot:xlabel='x [m]', ylabel='y [m]'>For a more complicated example, here is the deviation from Keplerian velocity around a planet embedded in a protoplanetary disc.
Deviation from Keplerian velocity around a planet: at the disc midplane (left), and 10 (middle) and 20 au (right) above the disc midplane. See here for details.
Extra quantities not written to the snapshot file are available:
>>> snap['angular_momentum']
array([ ... ]) <Unit('kilogram * meter ** 2 / second')>You can generate radial profiles on the snapshot. For example, to calculate the scale height in a disc:
>>> prof = plonk.load_profile(snap)
>>> prof['scale_height']
array([ ... ]) <Unit('meter')>Physical units of array quantities and other properties allow for unit conversion:
>>> pos = snap['position'][0]
>>> pos
array([-3.69505001e+12, 7.42032967e+12, -7.45096980e+11]) <Unit('meter')>
>>> pos.to('au')
array([-24.6998837 , 49.60184016, -4.98066567]) <Unit('astronomical_unit')>You can get a subset of particles as a SubSnap.
>>> subsnap = snap[:1000]
>>> subsnap = snap[snap['x'] > 0]
>>> subsnap = snap.family('gas')For further usage, see documentation. The code is internally documented with docstrings. Try, for example, help(plonk.Snap) or help(plonk.load_snap).
You can install Plonk via the package manager Conda from conda-forge.
conda install plonk --channel conda-forgeThis will install the required dependencies.
Note: You can simply use conda install plonk if you add the conda-forge channel with conda config --add channels conda-forge. I also recommend strictly using conda-forge which you can do with conda config --set channel_priority true. Both of these commands modify the Conda configuration file ~/.condarc.
You can also install Plonk from PyPI via pip.
python -m pip install plonkThis should install the required dependencies.
You can install Plonk from source as follows.
# clone via HTTPS
git clone https://github.com/dmentipl/plonk.git
# or clone via SSH
git clone [email protected]:dmentipl/plonk
cd plonk
python -m pip install -e .This assumes you have already installed the dependencies. One way to do this is by setting up a conda environment. The environment.yml file provided sets up a conda environment "plonk" for using or developing Plonk.
conda env create --file environment.yml
conda activate plonkPython 3.6+ with h5py, matplotlib, numba, numpy, pandas, pint, scipy, toml. Installing Plonk with conda or pip will install these dependencies.
If you need help, please try the following:
- Check the documentation.
- Check the built-in help, e.g.
help(plonk.load_snap). - Raise an issue, as a bug report or feature request, using the issue tracker.
Please don't be afraid to raise an issue. Even if your issue is not a bug, it's nice to have the question and answer available in a public forum so we can all learn from it together.
If you don't get an immediate response, please be patient. Plonk is maintained by one person, @dmentipl.
All types of contributions are welcome from all types of people with different skill levels.
Thank you for considering contributing to Plonk. There are many ways to contribute:
- If you find any bugs or cannot work out how to do something, please file a bug report in the issue tracker. Even if the issue is not a bug it may be that there is a lack of documentation.
- If you have any suggestions for new features, please raise a feature request in the issue tracker.
- If you use Plonk to do anything please consider contributing to the examples section in particular, or any other section, of the documentation.
- If you would like to contribute code, firstly thank you! We take code contributions via pull request.
See CONTRIBUTING.md for detailed guidelines on how to contribute.
If you use Plonk in a scientific publication, please cite the paper published in JOSS.
Plonk: Smoothed particle hydrodynamics analysis and visualization with Python
A BibTeX entry is available in CITATION.bib
If you use the interpolation to pixel grid component of Plonk please cite the Splash paper. You should also consider citing any other scientific software packages that you use.
The change log is available in CHANGELOG.md