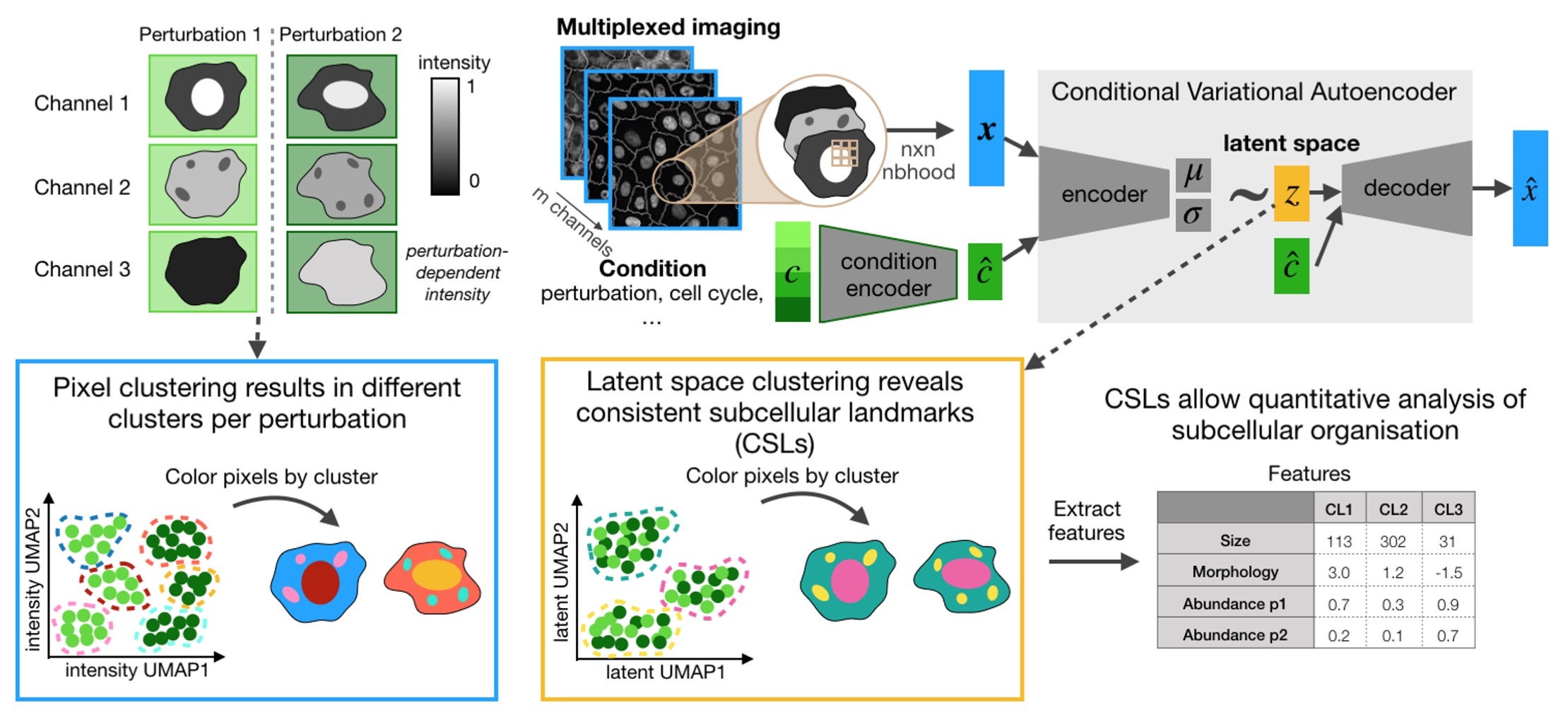
CAMPA is a framework for quantiative analysis of subcellular multi-channel imaging data. It consists of a workflow that generates consistent subcellular landmarks (CSLs) using conditional Variational Autoencoders (cVAE). The output of the CAMPA workflow is an anndata object that contains interpretable per-cell features summarizing the molecular composition and spatial arrangement of CSLs inside each cell.
Visit our documentation for installation and usage examples.
Please see our preprint "Learning consistent subcellular landmarks to quantify changes in multiplexed protein maps" (Spitzer, Berry et al. (2022)) to learn more.
CAMPA was developed for Python 3.9 and can be installed by running:
pip install campa
We are happy about any contributions! Before you start, check out our contributing guide.