The goal of healthyR.ts is to provide a consistent verb framework for
performing time series analysis and forecasting on both administrative
and clinical hospital data.
You can install the released version of healthyR.ts from CRAN with:
install.packages("healthyR.ts")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("spsanderson/healthyR.ts")This is a basic example which shows you how to generate random walk data.
library(healthyR.ts)
library(ggplot2)
df <- ts_random_walk()
head(df)
#> # A tibble: 6 × 4
#> run x y cum_y
#> <dbl> <dbl> <dbl> <dbl>
#> 1 1 1 0.0541 1054.
#> 2 1 2 -0.143 904.
#> 3 1 3 -0.0285 878.
#> 4 1 4 0.245 1093.
#> 5 1 5 0.0658 1165.
#> 6 1 6 0.00266 1168.Now that the data has been generated, lets take a look at it.
df %>%
ggplot(
mapping = aes(
x = x
, y = cum_y
, color = factor(run)
, group = factor(run)
)
) +
geom_line(alpha = 0.8) +
ts_random_walk_ggplot_layers(df)That is still pretty noisy, so lets see this in a different way. Lets clear this up a bit to make it easier to see the full range of the possible volatility of the random walks.
library(dplyr)
library(ggplot2)
df %>%
group_by(x) %>%
summarise(
min_y = min(cum_y),
max_y = max(cum_y)
) %>%
ggplot(
aes(x = x)
) +
geom_line(aes(y = max_y), color = "steelblue") +
geom_line(aes(y = min_y), color = "firebrick") +
geom_ribbon(aes(ymin = min_y, ymax = max_y), alpha = 0.2) +
ts_random_walk_ggplot_layers(df)This package comes with a wide variety of functions from Data Generators
to Statistics functions. The function ts_random_walk() in the above
example is a Data Generator.
Let’s take a look at a plotting function.
data_tbl <- data.frame(
date_col = seq.Date(
from = as.Date("2020-01-01"),
to = as.Date("2022-06-01"),
length.out = 365*2 + 180
),
value = rnorm(365*2+180, mean = 100)
)
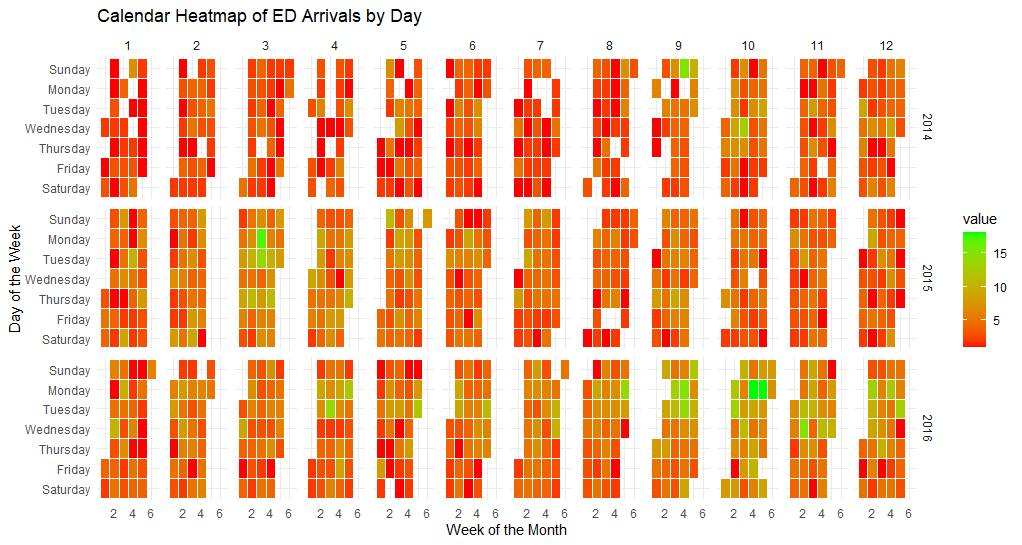
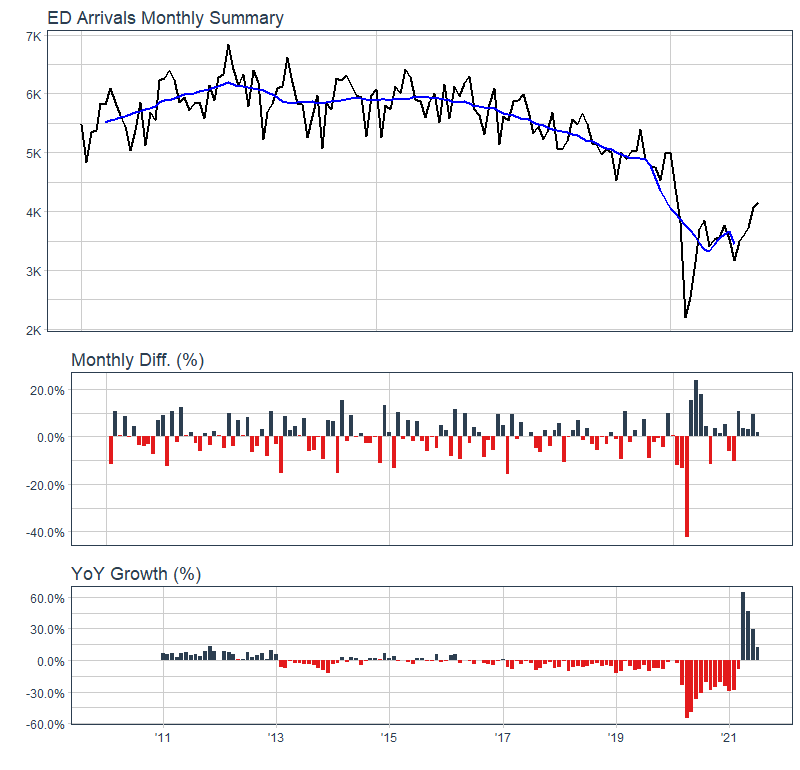
ts_calendar_heatmap_plot(
.data = data_tbl
, .date_col = date_col
, .value_col = value
, .interactive = FALSE
)Time Series Clustering via Features:
data_tbl <- ts_to_tbl(AirPassengers) %>%
mutate(group_id = rep(1:12, 12))
output <- ts_feature_cluster(
.data = data_tbl,
.date_col = date_col,
.value_col = value,
group_id,
.features = c("acf_features","entropy"),
.scale = TRUE,
.prefix = "ts_",
.centers = 3
)
ts_feature_cluster_plot(
.data = output,
.date_col = date_col,
.value_col = value,
.center = 2,
group_id
)Time to/from Event Analysis
library(dplyr)
df <- ts_to_tbl(AirPassengers) %>% select(-index)
ts_time_event_analysis_tbl(
.data = df,
.horizon = 6,
.date_col = date_col,
.value_col = value,
.direction = "both"
) %>%
ts_event_analysis_plot()ts_time_event_analysis_tbl(
.data = df,
.horizon = 6,
.date_col = date_col,
.value_col = value,
.direction = "both"
) %>%
ts_event_analysis_plot(.plot_type = "individual")ARIMA Simulators
output <- ts_arima_simulator()
output$plots$static_plotAutomatic Workflows which can be thought of as Boiler Plate Time Series modeling. This is in it’s infancy in this package.
| Auto Workflows | Boilerplate Workflow |
|---|---|
| ts_auto_arima() | Boilerplate Workflow |
| ts_auto_arima_xgboost() | Boilerplate Workflow |
| ts_auto_croston() | Boilerplate Workflow |
| ts_auto_exp_smoothing() | Boilerplate Workflow |
| ts_auto_glmnet() | Boilerplate Workflow |
| ts_auto_lm() | Boilerplate Workflow |
| ts_auto_mars() | Boilerplate Workflow |
| ts_auto_nnetar() | Boilerplate Workflow |
| ts_auto_prophet_boost() | Boilerplate Workflow |
| ts_auto_prophet_reg() | Boilerplate Workflow |
| ts_auto_smooth_es() | Boilerplate Workflow |
| ts_auto_svm_poly() | Boilerplate Workflow |
| ts_auto_svm_rbf() | Boilerplate Workflow |
| ts_auto_theta() | Boilerplate Workflow |
| ts_auto_xgboost() | Boilerplate Workflow |
This is just a start of what is in this package!