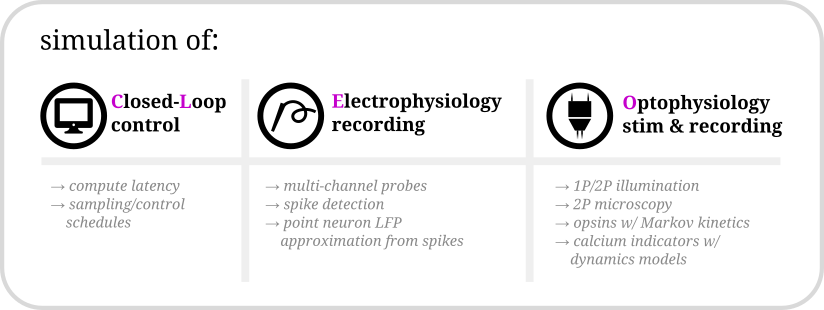
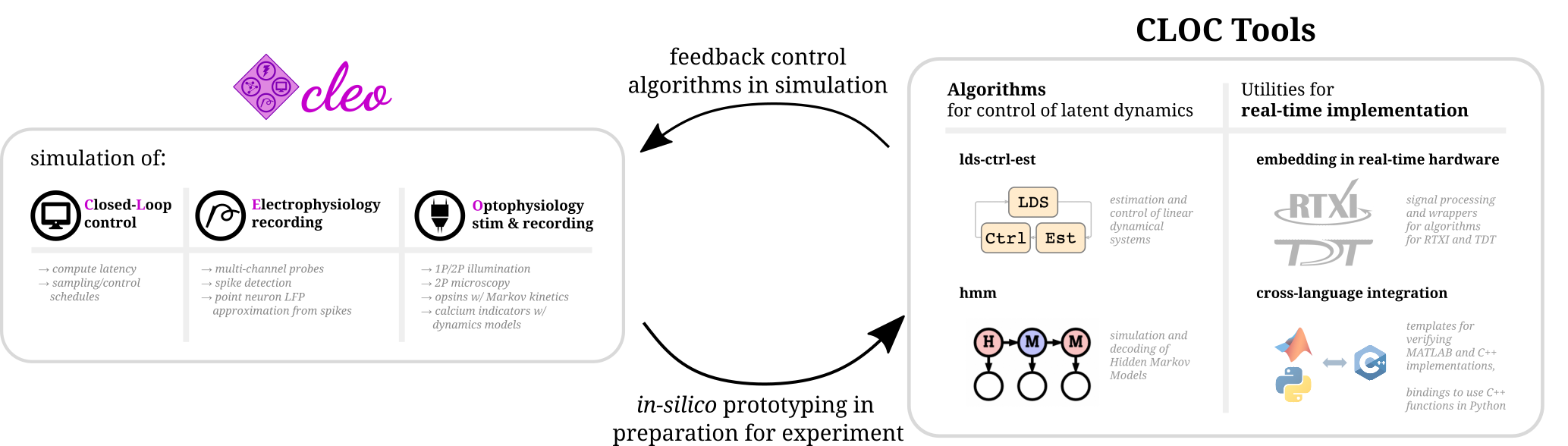
Hello there! Cleo has the goal of bridging theory and experiment for mesoscale neuroscience, facilitating electrode recording, optogenetic stimulation, and closed-loop experiments (e.g., real-time input and output processing) with the Brian 2 spiking neural network simulator. We hope users will find these components useful for prototyping experiments, innovating methods, and testing observations about a hypotheses in silico, incorporating into spiking neural network models laboratory techniques ranging from passive observation to complex model-based feedback control. Cleo also serves as an extensible, modular base for developing additional recording and stimulation modules for Brian simulations.
This package was developed by Kyle Johnsen and Nathan Cruzado under the direction of Chris Rozell at Georgia Institute of Technology. See the preprint here.
Cleo allows for flexible I/O processing in real time, enabling the simulation of closed-loop experiments such as event-triggered or feedback control. The user can also add latency to the stimulation to study the effects of computation delays.
Cleo provides functions for configuring electrode arrays and placing them in arbitrary locations in the simulation. The user can then specify parameters for probabilistic spike detection or a spike-based LFP approximation developed by Teleńczuk et al., 2020.
By modeling light propagation and opsins, Cleo enables users to flexibly add photostimulation to their model. Both a four-state Markov state model of opsin kinetics is available, as well as a minimal proportional current option for compatibility with simple neuron models. Cleo also accounts for opsin action spectra to model the effects of multi-light/wavelength/opsin crosstalk and heterogeneous expression. Parameters are for multiple opsins, and blue optic fiber (1P) and infrared spot (for 2P) illumination.
Users can also inject a microscope into their model, selecting neurons on the specified plane of imaging or elsewhere, with signal and noise strength determined by indicator expression levels and position with respect to the focal plane. The calcium indicator model of Song et al., 2021 is implemented, with parameters included for GCaMP6 variants.
Just use pip to install—the name on PyPI is cleosim:
pip install cleosimThen head to the overview section of the documentation for a more detailed discussion of motivation, structure, and basic usage.
Those using Cleo to simulate closed-loop control experiments may be interested in software developed for the execution of real-time, in-vivo experiments. Developed by members of Chris Rozell's and Garrett Stanley's labs at Georgia Tech, the CLOCTools repository can serve these users in two ways:
- By providing utilities and interfaces with experimental platforms for moving from simulation to reality.
- By providing performant control and estimation algorithms for feedback control.
Although Cleo enables closed-loop manipulation of network simulations, it does not include any advanced control algorithms itself.
The
ldsCtrlEstlibrary implements adaptive linear dynamical system-based control while thehmmlibrary can generate and decode systems with discrete latent states and observations.
Cleo: A testbed for bridging model and experiment by simulating closed-loop stimulation, electrode recording, and optophysiology
K.A. Johnsen, N.A. Cruzado, Z.C. Menard, A.A. Willats, A.S. Charles, and C.J. Rozell. bioRxiv, 2023.
CLOC Tools: A Library of Tools for Closed-Loop Neuroscience
A.A. Willats, M.F. Bolus, K.A. Johnsen, G.B. Stanley, and C.J. Rozell. In prep, 2023.
State-Aware Control of Switching Neural Dynamics
A.A. Willats, M.F. Bolus, C.J. Whitmire, G.B. Stanley, and C.J. Rozell. In prep, 2023.
Closed-Loop Identifiability in Neural Circuits
A. Willats, M. O'Shaughnessy, and C. Rozell. In prep, 2023.
State-space optimal feedback control of optogenetically driven neural activity
M.F. Bolus, A.A. Willats, C.J. Rozell and G.B. Stanley. Journal of Neural Engineering, 18(3), pp. 036006, March 2021.
Design strategies for dynamic closed-loop optogenetic neurocontrol in vivo
M.F. Bolus, A.A. Willats, C.J. Whitmire, C.J. Rozell and G.B. Stanley. Journal of Neural Engineering, 15(2), pp. 026011, January 2018.