Repository containing ZETA functions and dependencies. For an example of how to use the code, check runExampleZETA.m in the /example/ subfolder. Your output should look like the .tif in the same directory.
Note on updates and maintenance: this repository will stay as it is for backward compatibility. We have, however, been working on a version of ZETA for calcium imaging time-series dF/F0 data, and also made some minor changes (including syntax) to the zeta-test. We update the (now legacy) getZeta.m only to fix an occasional programming bug now and then, but getZeta.m and will not change otherwise. If you don't want to be bothered with updates and are happy with getZeta, you can stick with this repository. If you wish to have potentially slightly better performance on the ZETA-test, or would like to try out the time-series ZETA, you can switch to this repository: https://github.com/JorritMontijn/zetatest
The article describing ZETA has been published in eLife: https://elifesciences.org/articles/71969
This repository contains three main functions:
- getZeta.m: Calculates the Zenith of Event-based Time-locked Anomalies (ZETA) for spike times of a single neuron. Outputs a p-value.
- getMultiScaleDeriv.m: Calculates instantaneous firing rates for trace-based data, such as spike-time/z-score combinations that underlie ZETA.
- getIFR.m: Wrapper function for getMultiScaleDeriv.m when the input data are spike times and event times. Use this as you would a PSTH function.
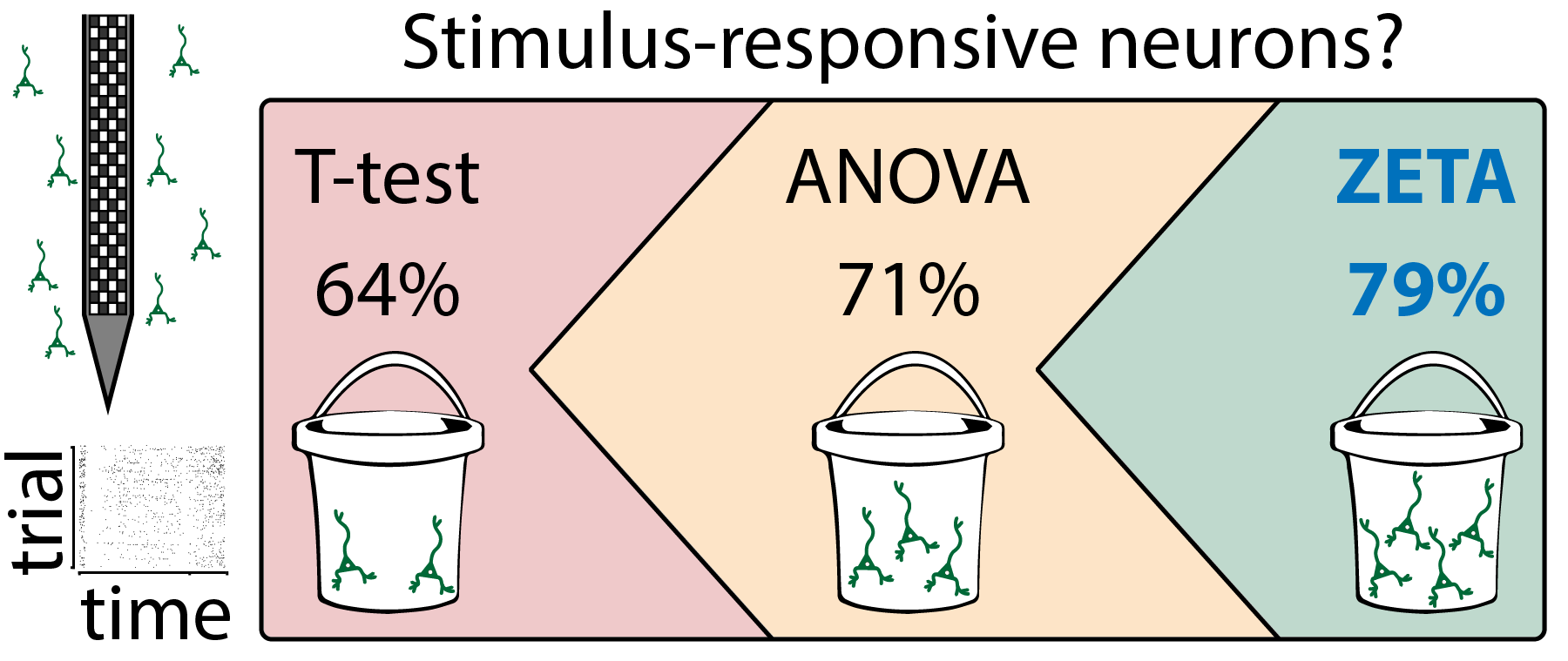
Rationale for ZETA
Neurophysiological studies depend on a reliable quantification of whether and when a neuron responds to stimulation, be it sensory, optogenetically or otherwise. However, current statistical analysis methods to determine a neuron’s responsiveness require arbitrary parameter choices, such as a binning size. This choice can change the results of the analysis, which invites bad statistical practice and reduces the replicability of analyses. Moreover, many methods, such as bin-wise t-tests, only detect classically mean-rate modulated cells. Especially with advent of techniques that yield increasingly large numbers of cells, such as Neuropixels recordings , it is important to use tests for cell-inclusion that require no manual curation. Here, we present the parameter-free ZETA-test, which outperforms common approaches, in the sense that it includes more cells at a similar false-positive rate. Finally, ZETA’s timescale-, parameter- and binning-free nature allowed us to implement a ZETA-derived algorithm (using multi-scale derivatives) to calculate peak onset and offset latencies in neuronal spike trains with theoretically unlimited temporal resolution.
Please send any questions or comments to j.montijn at nin.knaw.nl.
Dependencies getZeta.m requires the following Mathworks toolboxes to work:
- Signal Processing Toolbox
- Image Processing Toolbox
- Statistics and Machine Learning Toolbox
- (Optional: Parallel Computing Toolbox to reduce computation time)