helske / walker Goto Github PK
View Code? Open in Web Editor NEWBayesian Generalized Linear Models with Time-Varying Coefficients
License: GNU General Public License v3.0
Bayesian Generalized Linear Models with Time-Varying Coefficients
License: GNU General Public License v3.0
Currently all the regression coefficients are modeled as time-varying. But sometimes we might want to fix some of the coefficients as time-invariant. This poses no problems in theory, but would need some modifications to the codes.
Hi,
Walker apparently depends on Stan which is New BSD licensed but we are likely to add a dependency on an Apache 2 lib. This impacts GPL2 licensed projects. The voting thread is at https://discourse.mc-stan.org/t/apache-2-library-inclusion-into-stan-math-general-policy-case-trip-wire-consensus-decision/10929
We are seeking comments up until midnight Sept 14th anywhere on the planet.
Per the 15.10.2018 update it says that:
Missing values in response variable are now supported.
However, this gives the error:
y <- c(1:10, NA)
fit <- walker(y ~ -1 +
rw1(~ 1, beta_prior = c(0, 1), sigma_prior = c(0, 1)),
sigma_y_prior = c(0, 1), iter = 10, refresh=0)
Error in walker(y ~ -1 + rw1(~1, beta_prior = c(0, 1), sigma_prior = c(0, :
Missing values in response are not (yet) allowed.
I'm attempting to implement a pure random walk model in which the underlying data generating process is described by where y is a proportion between [0, 1]. The underlying state, y, is observed through counts of successes and failures using a logit link.
The problem is that when I forecast from model fit with walker_glm(distribution = "binomial"), the prediction intervals behave strangely. Specifically, there is a very large excess of predictions at exactly 0.5 (corresponding to zero on the logit scale). The remainder of the posterior predictive distribution aligns with my expectations. We can visualize this problem in the posterior predictive intervals,
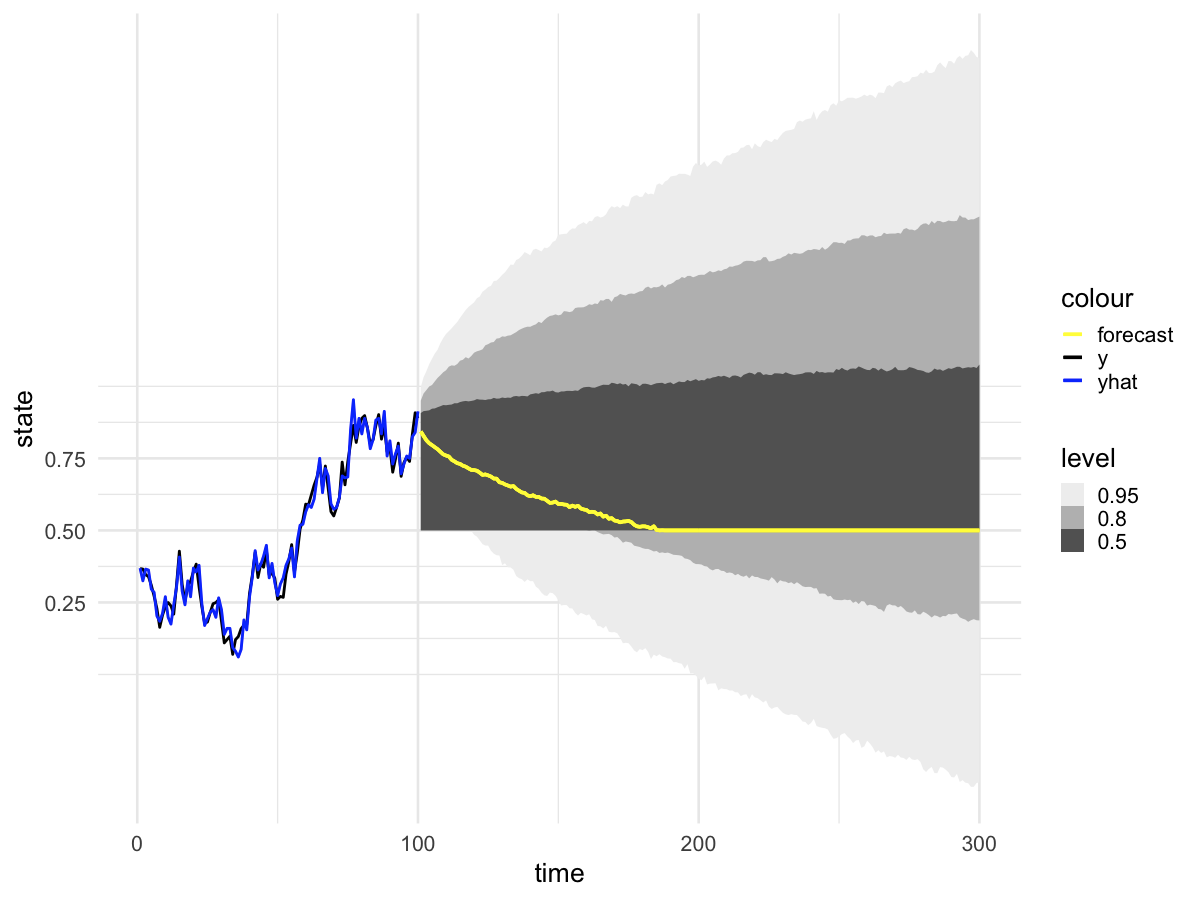
and in the posterior predictions at time=101
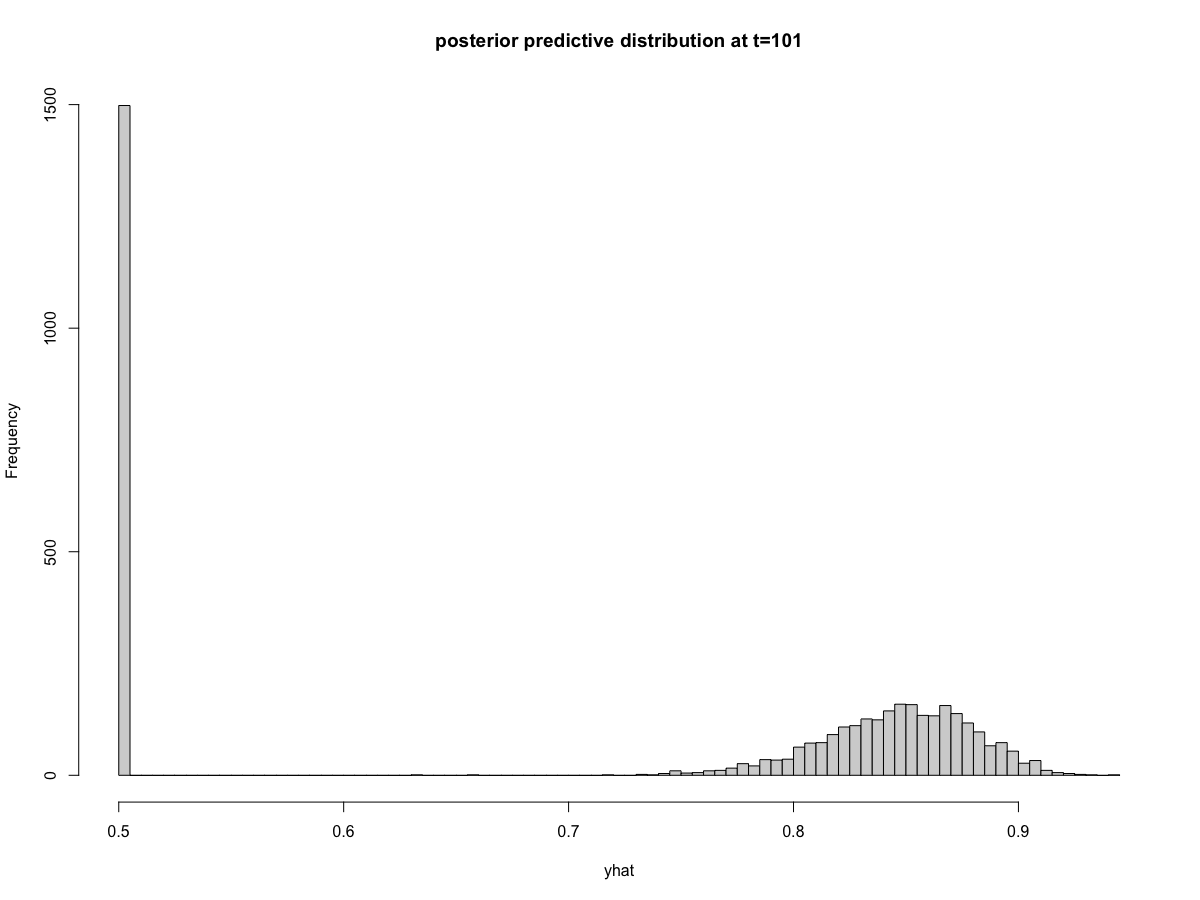
I noticed that all timepoints appear to have the same exact number of posterior predictions that equal 0.5 (again, this is the same as saying zero on the logit scale). You can see this in the histograms above where 1498 draws equal 0.5 at both t=101 and t=250 (the exact count will vary depending on the random seed). The reprex below provides code that shows this is true across all time points. I can't think of a mechanism that would cause consistency of posterior draws across every time point.
We can simulate nt=100 time steps of the true data y, the number of observed successes succ out of nobs trials, and the estimated state on the probability scale yhat as:
# preliminaries
library(walker)
library(tidyverse)
library(ggedit) # used later to remove count layer from plot_predict()
## simulate data generating process
## (a random walk proportion, estimated from observed successes)
# parameters
sigma <- 0.25 # standard deviation of the 'step size' on logit scale
# sample sizes
nt <- 100 # duration of observed timeseries
nobs <- 300 # total number of observations of events with probability 'y'
# generate latent process data
set.seed(32101)
rw <- cumsum(rnorm(nt, 0, sigma)) # random walk process on logit scale
y <- plogis(rw) # timeseries of _true_ Pr(success)
# simulate observations of latent process
succ <- rbinom(nt, nobs, y) # timeseries of observed successes
yhat <- succ / nobs # timeseries of _estimated_ Pr(success)
plot.ts(y, ylim = c(0, 1)) # true Pr(success)
lines(yhat, col = "blue") # estimated Pr(success)
dat <- data.frame(time = 1:nt, rw, y, succ, nobs, yhat)
We can then fit the model using walker_glm() with a very weak prior on the value of yhat at time=0, and confirm that the parameters are correctly recovered:
## pick a flat prior for the observed proportions
plot(density(plogis(rnorm(1e6, mean = 0, sd = 1.45))))
## fit a pure random walk model to the data (takes ~90s)
rwfit <- walker_glm(succ ~ 0 + # don't fit an overall intercept
rw1(~ 0, # don't fit a time-varying intercept or covariates
beta = c(0, 1.45), # prior: Normal(0,1.45) on observation at t0 (logit scale)
sigma = c(0, 2)), # prior: Half-Normal(0,2) on 'sigma'
u = dat$nobs, # total number of plants counted
distribution = "binomial", # model ratio from count data
data = dat, iter = 2000, warmup = 1000, chains = 4, cores = 4)
## check convergence
rwfit
pp_check(rwfit)
## check that sigma is correctly recovered
sigma_est <- unlist(lapply(rwfit[["stanfit"]]@sim[["samples"]], function(x) x[["sigma_rw1[1]"]]))
hist(sigma_est)
abline(v = sigma, col = "blue")
Finally, make the forecast for times 101 to 300:
## forecast the model from times 101:300
ndat <- data.frame(time = 1:length(101:300))
rwfc <- predict(rwfit, newdata = ndat, u = nobs)
## plot forecast using internal walker function
plot_predict(rwfc) %>%
remove_geom("line", 2)
## re-plot with tidybayes to show multiple posterior intervals
plotdat <- as.data.frame(rwfc[["y_new"]]) %>%
rownames_to_column(var = "time") %>%
pivot_longer(-time, names_to = ".iter", values_to = "yhat") %>%
mutate_if(is.character, as.integer)
ggplot(plotdat, aes(x = time, y = yhat))+
stat_lineribbon(aes(color = "forecast"))+
geom_line(data = dat, aes(y = y, color = "y"), size = 1)+
geom_line(data = dat, aes(y = yhat, color = "yhat"), size = 1)+
scale_y_continuous(name = "state", trans = scales::logit_trans())+
scale_color_manual(values = c("yellow", "black", "blue"))+
scale_fill_brewer(palette = "Greys")+
theme_minimal(base_size = 20)
## look at forecast posterior at t=101
plotdat %>%
filter(time == 101) %>%
pull(yhat) %>%
hist(100, main = "posterior predictive distribution at t=101",
xlab = "yhat")
## look at forecast posterior at t=250
plotdat %>%
filter(time == 250) %>%
pull(yhat) %>%
hist(100, main = "posterior predictive distribution at t=250",
xlab = "yhat")
# count the number of yhat == 0.5 at each time point
plotdat %>%
mutate(test = yhat==0.5) %>%
group_by(.$time) %>%
summarize(count = sum(test))
Thank you, @helske, for your work on this package to make state space modeling intuitive and efficient.
Thanks for the update to allowing NAs. I'm running some models where I'm interested in what would have happened if my covariates had taken on different values in the past. I'm OK with assuming that everything else would have been the same except for the effect of the covariates. I've resorted to adding in an extra data parameter (counterfact_xreg_rw) in the data block and y_counterfact[N] in generated quantities:
// replicated data from posterior predictive distribution
for(t in 1:n) {
y_counterfact[t] = xbeta[t] + dot_product(counterfact_xreg_rw[, t] * beta_rw[, t]);
y_fit[t] = xbeta[t] + dot_product(xreg_rw[, t], beta_rw[, t]);
y_rep[t] = normal_rng(y_fit[t], gamma_y[t] * sigma_y);
}
I'm wondering if this could be included in the predict function with an optional parameter pass to indicate that this is for counterfactual predictions during the estimation time period?
Got an email from CRAN, address sanitizer gives an error:
* using log directory ‘/data/gannet/ripley/R/packages/tests-gcc-SAN/walker.Rcheck’
* using R Under development (unstable) (2021-01-25 r79883)
* using platform: x86_64-pc-linux-gnu (64-bit)
* using session charset: UTF-8
* using option ‘--no-stop-on-test-error’
* checking for file ‘walker/DESCRIPTION’ ... OK
* checking extension type ... Package
* this is package ‘walker’ version ‘1.0.1’
* package encoding: UTF-8
* checking package dependencies ... OK
* checking if this is a source package ... OK
* checking if there is a namespace ... OK
* checking for hidden files and directories ... OK
* checking for portable file names ... OK
* checking whether package ‘walker’ can be installed ... [61m/61m] WARNING
Found the following significant warnings:
Warning: namespace ‘DBI’ is not available and has been replaced
See ‘/data/gannet/ripley/R/packages/tests-gcc-SAN/walker.Rcheck/00install.out’ for details.
* checking package directory ... OK
* checking whether the package can be loaded ... OK
* checking whether the package can be loaded with stated dependencies ... OK
* checking whether the package can be unloaded cleanly ... OK
* checking whether the namespace can be loaded with stated dependencies ... OK
* checking whether the namespace can be unloaded cleanly ... OK
* checking loading without being on the library search path ... OK
* checking compiled code ... OK
* checking installed files from ‘inst/doc’ ... OK
* checking files in ‘vignettes’ ... OK
* checking examples ... [87s/88s] OK
* checking tests ... [22s/22s] OK
Running ‘test_all.R’ [21s/21s]
* checking package vignettes in ‘inst/doc’ ... OK
* checking re-building of vignette outputs ... [15m/15m] WARNING
Error(s) in re-building vignettes:
--- re-building ‘walker.Rmd’ using rmarkdown
=================================================================
==799014==ERROR: AddressSanitizer: heap-buffer-overflow on address 0x6020013a9e38 at pc 0x7f6889ef739a bp 0x7ffc79d59b00 sp 0x7ffc79d592a8
READ of size 4000 at 0x6020013a9e38 thread T0
#0 0x7f6889ef7399 in memmove (/lib64/libasan.so.6+0x3a399)
#1 0x7f685fc2bb46 in double* std::__copy_move<false, true, std::random_access_iterator_tag>::__copy_m<double>(double const*, double const*, double*) /usr/include/c++/10/bits/stl_algobase.h:426
#2 0x7f685fc2bb46 in double* std::__copy_move_a2<false, double*, double*>(double*, double*, double*) /usr/include/c++/10/bits/stl_algobase.h:472
#3 0x7f685fc2bb46 in double* std::__copy_move_a1<false, double*, double*>(double*, double*, double*) /usr/include/c++/10/bits/stl_algobase.h:506
#4 0x7f685fc2bb46 in double* std::__copy_move_a<false, __gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, double*>(__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, __gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, double*) /usr/include/c++/10/bits/stl_algobase.h:513
#5 0x7f685fc2bb46 in double* std::copy<__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, double*>(__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, __gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, double*) /usr/include/c++/10/bits/stl_algobase.h:569
#6 0x7f685fc2bb46 in void std::vector<double, std::allocator<double> >::_M_assign_aux<__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > > >(__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, __gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, std::forward_iterator_tag) /usr/include/c++/10/bits/vector.tcc:321
#7 0x7f685fc2ed5c in void std::vector<double, std::allocator<double> >::_M_assign_dispatch<__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > > >(__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, __gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, std::__false_type) /usr/include/c++/10/bits/stl_vector.h:1628
#8 0x7f685fc2ed5c in void std::vector<double, std::allocator<double> >::assign<__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, void>(__gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >, __gnu_cxx::__normal_iterator<double*, std::vector<double, std::allocator<double> > >) /usr/include/c++/10/bits/stl_vector.h:769
#9 0x7f685fc2ed5c in split_potential_scale_reduction /tmp/Rtmp5Oq3an/R.INSTALL327d7a20b63ed9/rstan/src/chains.cpp:568
#10 0x57d4a7 in R_doDotCall /data/gannet/ripley/R/svn/R-devel/src/main/dotcode.c:601
#11 0x639251 in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7671
#12 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#13 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#14 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#15 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#16 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#17 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#18 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#19 0x67decd in R_forceAndCall /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1964
#20 0x44c88a in do_lapply /data/gannet/ripley/R/svn/R-devel/src/main/apply.c:70
#21 0x71bba9 in do_internal /data/gannet/ripley/R/svn/R-devel/src/main/names.c:1397
#22 0x623b9c in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7135
#23 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#24 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#25 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#26 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#27 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#28 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#29 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#30 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#31 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#32 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#33 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#34 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#35 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#36 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#37 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#38 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#39 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#40 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#41 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#42 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#43 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#44 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#45 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#46 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#47 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#48 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#49 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#50 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#51 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#52 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#53 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#54 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#55 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#56 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#57 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#58 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#59 0x6786e6 in R_execMethod /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:2073
#60 0x7f687d1b8429 in R_dispatchGeneric /data/gannet/ripley/R/svn/R-devel/src/library/methods/src/methods_list_dispatch.c:1140
#61 0x728310 in do_standardGeneric /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:1285
#62 0x6716ba in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:834
#63 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#64 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#65 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#66 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#67 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#68 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#69 0x72155e in dispatchMethod /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:436
#70 0x72205f in Rf_usemethod /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:476
#71 0x722b93 in do_usemethod /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:565
#72 0x623b9c in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7135
#73 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#74 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#75 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#76 0x670ecf in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:850
#77 0x67fe27 in do_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:3344
#78 0x6288eb in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7115
#79 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#80 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#81 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#82 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#83 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#84 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#85 0x670c13 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:750
#86 0x68167c in do_withVisible /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:3396
#87 0x71bba9 in do_internal /data/gannet/ripley/R/svn/R-devel/src/main/names.c:1397
#88 0x623b9c in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7135
#89 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#90 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#91 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#92 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#93 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#94 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#95 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#96 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#97 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#98 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#99 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#100 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#101 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#102 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#103 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#104 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#105 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#106 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#107 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#108 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#109 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#110 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#111 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#112 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#113 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#114 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#115 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#116 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#117 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#118 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#119 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#120 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#121 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#122 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#123 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#124 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#125 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#126 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#127 0x670ecf in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:850
#128 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#129 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#130 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#131 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#132 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#133 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#134 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#135 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#136 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#137 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#138 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#139 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#140 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#141 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#142 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#143 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#144 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#145 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#146 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#147 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#148 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#149 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#150 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#151 0x72155e in dispatchMethod /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:436
#152 0x72205f in Rf_usemethod /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:476
#153 0x722b93 in do_usemethod /data/gannet/ripley/R/svn/R-devel/src/main/objects.c:565
#154 0x623b9c in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7135
#155 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#156 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#157 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#158 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#159 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#160 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#161 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#162 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#163 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#164 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#165 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#166 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#167 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#168 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#169 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#170 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#171 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#172 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#173 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#174 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#175 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#176 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#177 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#178 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#179 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#180 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#181 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#182 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#183 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#184 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#185 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#186 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#187 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#188 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#189 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#190 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#191 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#192 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#193 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#194 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#195 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#196 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#197 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#198 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#199 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#200 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#201 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#202 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#203 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#204 0x672652 in forcePromise /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:555
#205 0x672de7 in FORCE_PROMISE /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5136
#206 0x672de7 in getvar /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:5177
#207 0x63727a in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:6867
#208 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#209 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#210 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#211 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#212 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#213 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#214 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#215 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#216 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#217 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#218 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#219 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#220 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#221 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#222 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#223 0x64704e in bcEval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:7083
#224 0x670607 in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:727
#225 0x6758f4 in R_execClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1897
#226 0x677d97 in Rf_applyClosure /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:1823
#227 0x670ecf in Rf_eval /data/gannet/ripley/R/svn/R-devel/src/main/eval.c:850
#228 0x6f0c0d in Rf_ReplIteration /data/gannet/ripley/R/svn/R-devel/src/main/main.c:264
#229 0x6f1258 in R_ReplConsole /data/gannet/ripley/R/svn/R-devel/src/main/main.c:314
#230 0x6f13a4 in run_Rmainloop /data/gannet/ripley/R/svn/R-devel/src/main/main.c:1113
#231 0x6f13f2 in Rf_mainloop /data/gannet/ripley/R/svn/R-devel/src/main/main.c:1120
#232 0x41b3c8 in main /data/gannet/ripley/R/svn/R-devel/src/main/Rmain.c:29
#233 0x7f6888877081 in __libc_start_main (/lib64/libc.so.6+0x27081)
#234 0x41db3d in _start (/data/gannet/ripley/R/gcc-SAN/bin/exec/R+0x41db3d)
0x6020013a9e38 is located 0 bytes to the right of 8-byte region [0x6020013a9e30,0x6020013a9e38)
allocated by thread T0 here:
#0 0x7f6889f6f067 in operator new(unsigned long) (/lib64/libasan.so.6+0xb2067)
#1 0x7f685fc2b97f in __gnu_cxx::new_allocator<double>::allocate(unsigned long, void const*) /usr/include/c++/10/ext/new_allocator.h:115
#2 0x7f685fc2b97f in std::allocator_traits<std::allocator<double> >::allocate(std::allocator<double>&, unsigned long) /usr/include/c++/10/bits/alloc_traits.h:460
#3 0x7f685fc2b97f in std::_Vector_base<double, std::allocator<double> >::_M_allocate(unsigned long) /usr/include/c++/10/bits/stl_vector.h:346
#4 0x7f685fc2b97f in double* std::vector<double, std::allocator<double> >::_M_allocate_and_copy<double*>(unsigned long, double*, double*) /usr/include/c++/10/bits/stl_vector.h:1511
#5 0x7f685fc2b97f in void std::vector<double, std::allocator<double> >::_M_assign_aux<double*>(double*, double*, std::forward_iterator_tag) /usr/include/c++/10/bits/vector.tcc:309
SUMMARY: AddressSanitizer: heap-buffer-overflow (/lib64/libasan.so.6+0x3a399) in memmove
Shadow bytes around the buggy address:
0x0c048026d370: fa fa fd fd fa fa fd fa fa fa fd fa fa fa fd fa
0x0c048026d380: fa fa fd fd fa fa fd fa fa fa fd fa fa fa fd fd
0x0c048026d390: fa fa fd fd fa fa fd fd fa fa 01 fa fa fa 01 fa
0x0c048026d3a0: fa fa 01 fa fa fa 01 fa fa fa 01 fa fa fa 01 fa
0x0c048026d3b0: fa fa fa fa fa fa fd fa fa fa fd fd fa fa fd fa
=>0x0c048026d3c0: fa fa fd fa fa fa 00[fa]fa fa 04 fa fa fa 04 fa
0x0c048026d3d0: fa fa fa fa fa fa fa fa fa fa fa fa fa fa fa fa
0x0c048026d3e0: fa fa fa fa fa fa fa fa fa fa fa fa fa fa fd fd
0x0c048026d3f0: fa fa 04 fa fa fa fd fd fa fa fd fa fa fa fd fa
0x0c048026d400: fa fa fa fa fa fa 01 fa fa fa 01 fa fa fa 01 fa
0x0c048026d410: fa fa 01 fa fa fa 01 fa fa fa 01 fa fa fa fa fa
Shadow byte legend (one shadow byte represents 8 application bytes):
Addressable: 00
Partially addressable: 01 02 03 04 05 06 07
Heap left redzone: fa
Freed heap region: fd
Stack left redzone: f1
Stack mid redzone: f2
Stack right redzone: f3
Stack after return: f5
Stack use after scope: f8
Global redzone: f9
Global init order: f6
Poisoned by user: f7
Container overflow: fc
Array cookie: ac
Intra object redzone: bb
ASan internal: fe
Left alloca redzone: ca
Right alloca redzone: cb
Shadow gap: cc
==799014==ABORTING
* DONE
Status: 2 WARNINGs
Seems to be related to rstan (no surprise as almost all the compiled code is generated by Stan). For some reason the sanitizer check on rhub does not work at the moment, so not sure if it is just an issue on CRAN side.
The unit test "stan side works" in tests.R is not working for me. Those double precision numbers you are testing against depend on the operating system, the compiler, and the version of Stan. So, I would instead just test that the output contains a valid stanfit object.
In
walker/inst/stan/walker_lm.stan
Line 134 in 392bafb
walker_lm code but is initialized to all 1s. It's not documented that you can pass this argument in. Just wondering what it does.Current implementation of the simulation smoother algorithm used in walker_glm is somewhat slow, as I am not taking account the fact that we are simulating multiple samples from the same model. The corrections are relatively straightforward, and should be implemented at some point. Essentially same code is available in bssm package as well, so it should be pretty easy to just convert those codes to Stan.
* using log directory ‘/data/gannet/ripley/R/packages/tests-clang-SAN/walker.Rcheck’
* using R Under development (unstable) (2017-07-12 r72910)
* using platform: x86_64-pc-linux-gnu (64-bit)
* using session charset: UTF-8
* using option ‘--no-stop-on-test-error’
* checking for file ‘walker/DESCRIPTION’ ... OK
* checking extension type ... Package
* this is package ‘walker’ version ‘0.2.0’
* checking package dependencies ... OK
* checking if this is a source package ... OK
* checking if there is a namespace ... OK
* checking for hidden files and directories ... OK
* checking for portable file names ... OK
* checking whether package ‘walker’ can be installed ... [11m/12m] OK
* checking package directory ... OK
* checking whether the package can be loaded ... OK
* checking whether the package can be loaded with stated dependencies ... OK
* checking whether the package can be unloaded cleanly ... OK
* checking whether the namespace can be loaded with stated dependencies ... OK
* checking whether the namespace can be unloaded cleanly ... OK
* checking loading without being on the library search path ... OK
* checking compiled code ... OK
* checking installed files from ‘inst/doc’ ... OK
* checking files in ‘vignettes’ ... OK
* checking examples ... [132s/135s] OK
* checking tests ... [26s/27s] OK
Running ‘test_all.R’ [25s/25s]
* checking package vignettes in ‘inst/doc’ ... OK
* checking re-building of vignette outputs ... [60m/61m] WARNING
Error in re-building vignettes:
...
/data/gannet/ripley/R/test-clang/RcppArmadillo/include/armadillo_bits/subview_meat.hpp:1189:23: runtime error: reference binding to null pointer of type 'const double'
SUMMARY: AddressSanitizer: undefined-behavior /data/gannet/ripley/R/test-clang/RcppArmadillo/include/armadillo_bits/subview_meat.hpp:1189:23 in
* DONE
Status: 1 WARNING
I get the following errors when I try to run the example code for walker_glm. The same error occurs for both examples and when I try to use my own data.
data("discoveries", package = "datasets")
out <- walker_glm(discoveries ~ -1 +
rw2(~ 1, beta_prior = c(0, 10), sigma_prior = c(0, 2), slope_prior = c(0, 2)),
distribution = "poisson", iter = 1000, chains = 1, refresh = 0)
[1] "Error in sampler$call_sampler(args_list[[i]]) : "
[2] " Exception: assign: Rows of left-hand-side (100) and rows of right-hand-side (2) must match in size (in 'model_walker_glm' at line 325)"
[1] "error occurred during calling the sampler; sampling not done"
Currently we return standard stanfit objects, but it would be good to have some helper functions for analyzing the output. Currently some manual work is needed in order to plot the coefficient series etc. which could be automated pretty easily.
Opening separate issue following discussion in #11...
The predict.walker_fit method returns values on both the link and response scales for models with non-Gaussian errors. plot_predict plots both, and this is not currently controllable by the user. This behavior is particularly problematic for distribution='logit' with high numbers of trials u. In these cases, the response scale is so much higher than the link scale (constrained between [0,1]) that variation on the link scale is completely flattened.
As a temporary workaround, ggedit::remove_geom() can be used to remove the undesired set of predictions. For an example of use, see the first post in #11.
This issue requests that an argument be added to plot_predict that allows users to specify whether they want the posterior predictions plotted on the 'link' or 'response' scale. I advocate setting 'link' as the default, as this corresponds to the state that is usually of interest when doing state space modeling. This default would also mirror the behavior of well-known packages like lme4 (see ?predict.merMod).
We are struggling to get a new version of StanHeaders onto CRAN and walker is one of several packages that we are having issues with.
First, walker has Makevars and Makevars.win files in src/ . Ordinarily, a package that uses rstantools like walker does should not have those files, since they get regenerated automatically by rstantools. That is why there is a comment at the top of them saying that they are autogenerated and should not be edited by hand. When those files are modified by hand, then rstantools does not overwrite them. However, that now means that the necessary changes to the build process are not picked up, which makes walker segfault when its namespace is loaded with the provisional StanHeaders. Just by deleting the src/Makevars and src/Makevars.win files, I was able to get walker to build and load, so I am not understanding why it was necessary to change them.
Second, after doing that, walker has some errors in its vignette and examples in situations where the provisional StanHeaders catches mistakes that are somehow not being caught with the StanHeaders / rstan that are on CRAN currently. For example, when you run
example(plot_prediction, package = "walker")
with the provisional StanHeaders, you get an exception
Exception: gamma_lpdf: Shape parameter is 0, but must be positive finite! (in 'model_walker_lm' at line 195)
that prevents the Markov chains from initializing. If you look at the list of data being passed to your Stan program,
List of 28
$ k_fixed : num 1
$ k_rw1 : num 0
$ k_rw2 : num 1
$ m : num 2
$ k : num 1
$ n : int 50
$ n_lfo : int 50
$ xreg_fixed : num [1:50, 1] 1 1 1 1 1 1 1 1 1 1 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:50] "1" "2" "3" "4" ...
.. ..$ : chr "(Intercept)"
..- attr(*, "assign")= int 0
$ xreg_rw : num [1, 1:50] 2.201 0.98 1.345 1.014 0.628 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr "x"
.. ..$ : chr [1:50] "1" "2" "3" "4" ...
$ y : Named num [1:50] 4.07 3.31 3.28 2.95 2.59 ...
..- attr(*, "names")= chr [1:50] "1" "2" "3" "4" ...
$ y_miss : int [1:50] 0 0 0 0 0 0 0 0 0 0 ...
$ sigma_y_shape : num 2
$ sigma_y_rate : num 0.01
$ beta_fixed_mean: num 0
$ beta_fixed_sd : num 10
$ beta_rw1_mean : num 0
$ beta_rw1_sd : num 0
$ beta_rw2_mean : num 0
$ beta_rw2_sd : num 10
$ sigma_rw1_shape: num 0
$ sigma_rw1_rate : num 0
$ sigma_rw2_shape: num 2
$ sigma_rw2_rate : num 1e-04
$ nu_mean : num 0
$ nu_sd : num 10
$ gamma_y : num [1:50] 1 1 1 1 1 1 1 1 1 1 ...
$ gamma_rw1 : num[0 , 1:50]
$ gamma_rw2 : num [1, 1:50] 1 1 1 1 1 1 1 1 1 1 ...
indeed both sigma_rw1_shape and sigma_rw1_rate are zero, which is not a valid gamma distribution. If you want an improper uniform prior over the positive real numbers (which is not a very good idea), then just omit the prior for a parameter that has been declared with lower = 0. But I think this is more likely a mistake, albeit one that Stan should have caught many versions ago.
We really need you to upload a new walker to CRAN that fixes these issues, but before you do, let me know so that I can test it with the provisional StanHeaders. Thanks!
Currently, all time-varying covariates should be included in one rw1 and rw2 calls, with identical priors. This can be problematic in some cases. It would be relatively straightforward to parse multiple rw1 calls during the formula manipulation and then combine them before sending stuff to Stan side.
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.