Robocrystallographer is a tool to generate text descriptions of crystal structures. Similar to how a real-life crystallographer would analyse a structure, robocrystallographer looks at the symmetry, local environment, and extended connectivity when generating a description. The package includes utilities for identifying molecule names, component orientations, heterostructure information, and more...
Robocrystallographer can be used from the command-line or from a python API. The package integrates with the Materials Project to for allow generation of structure descriptions directly from Materials Project ids. For example, to generate the description of SnO2 (mp-856), one can simply run:
robocrys mp-856Alternatively, a structure file can be specified in place of a Materials Project id. Robocrystallographer supports the same file formats as pymatgen, including the Crystallographic Information Format (CIF), and common electronic structure package formats such as POSCAR files. More information can be found on the command-line interface page.
The two core classes in robocrystallographer are:
StructureCondenser: to condense the structure into an descriptive JSON representation.StructureDescriber: to turn the condensed structure into a text description.
A minimal working example for generating text descriptions is simply:
from pymatgen import Structure
from robocrys import StructureCondenser, StructureDescriber
structure = Structure.from_file("my_structure.cif") # other file formats also supported
# alternatively, uncomment the lines below to use the MPRester object
# to fetch structures from the Materials Project database
# from mp_api.client import MPRester
# structure = MPRester(api_key=None).get_structure_by_material_id("mp-856")
condenser = StructureCondenser()
describer = StructureDescriber()
condensed_structure = condenser.condense_structure(structure)
description = describer.describe(condensed_structure)Where structure is a pymatgen Structure object. Both classes have many
options for customising the output of the structure
descriptions. More information is provided in the
module documentation.
The format of the intermediate JSON representation is detailed on the condensed structure format page.
An example of the output generated by robocrystallographer for SnO2 (mp-856) is given below:
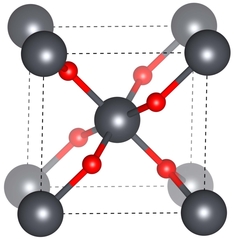
SnO2 is Rutile structured and crystallizes in the tetragonal P4_2/mnm space group. The structure is three-dimensional. Sn(1) is bonded to six equivalent O(1) atoms to form a mixture of edge and corner-sharing SnO6 octahedra. The corner-sharing octahedral tilt angles are 51°. All Sn(1)–O(1) bond lengths are 2.09 Å. O(1) is bonded in a trigonal planar geometry to three equivalent Sn(1) atoms.
Please considering citing the following publication if using robocrystallographer in your work:
Ganose, A., & Jain, A. (2019). Robocrystallographer: Automated crystal structure text descriptions and analysis. MRS Communications, 9(3), 874-881. https://doi.org/10.1557/mrc.2019.94
Robocrystallographer can be installed using pip:
pip install robocrysRobocrystallographer requires Python 3.7+. The OpenBabel package is required to determine molecule names. This is an optional requirement but its use is recommended for best results. If you are using the Conda package management system, OpenBabel can be installed using:
conda install -c conda-forge openbabelTrack changes to robocrystallographer through the Changelog.
Robocrystallographer is in early development but we still welcome your contributions. Please read our contribution guidelines for more information. We maintain a list of all contributors here.
Robocrystallographer is released under a modified BSD license; the full text can be found here.
Logo by Somewan from the Noun Project.