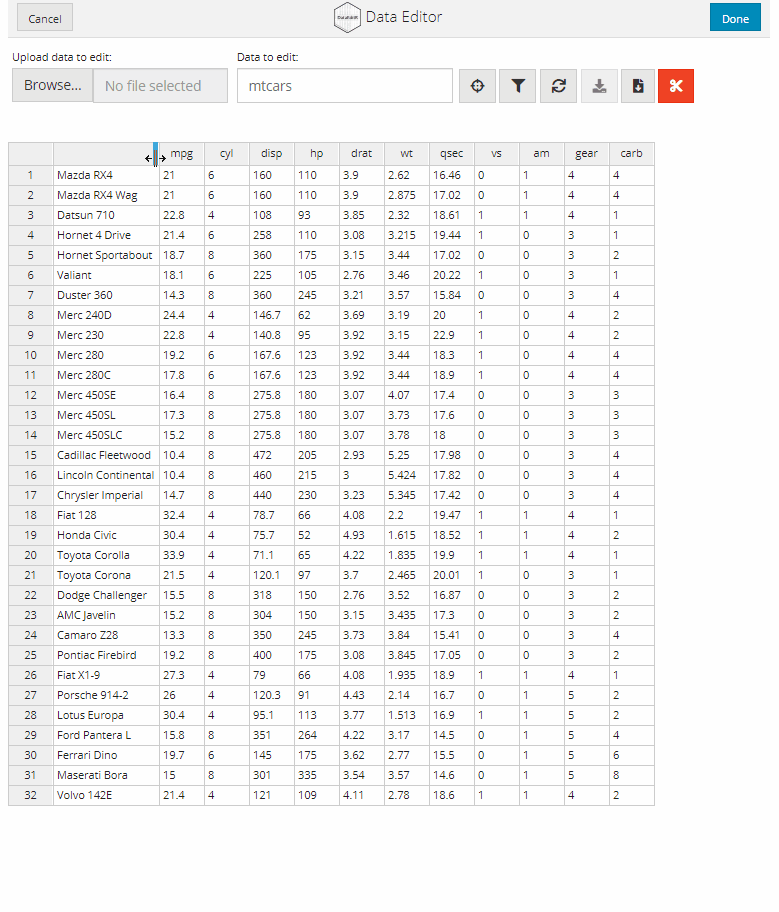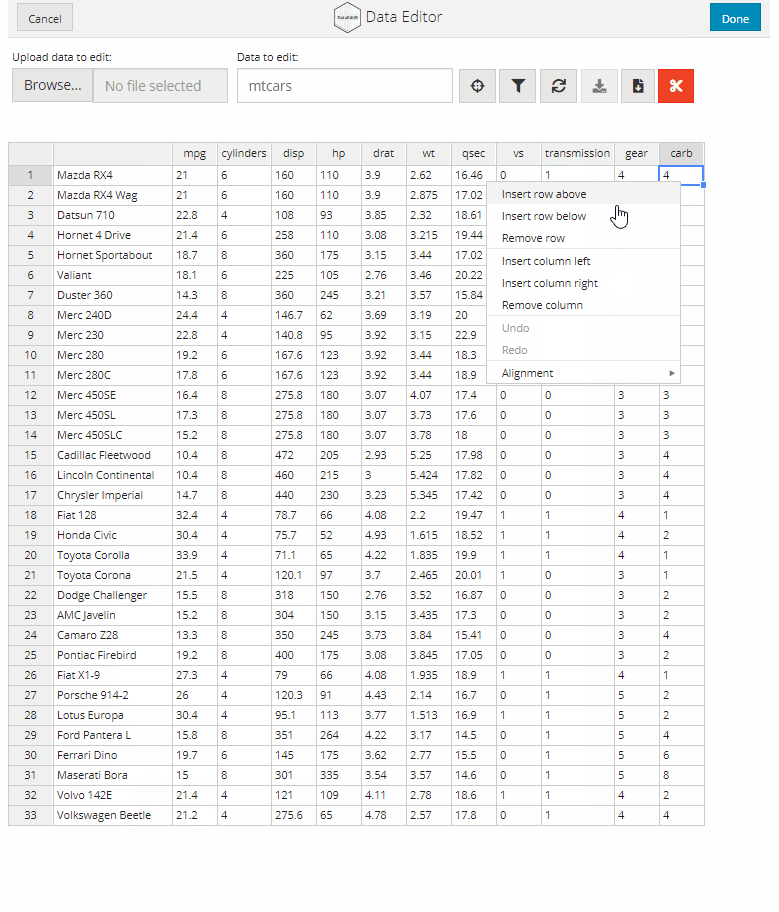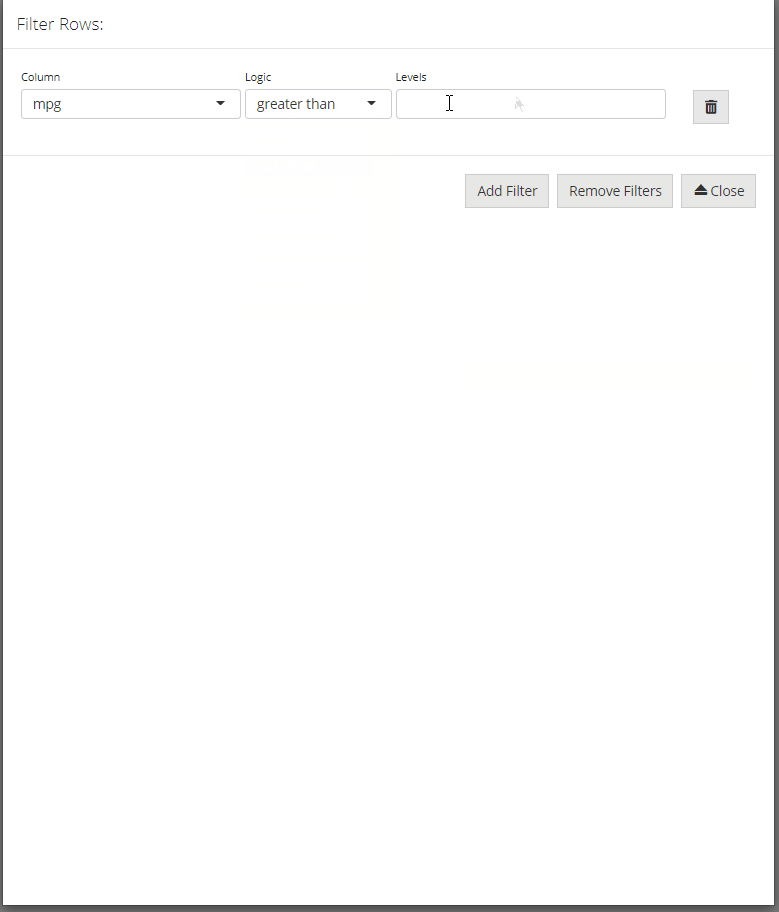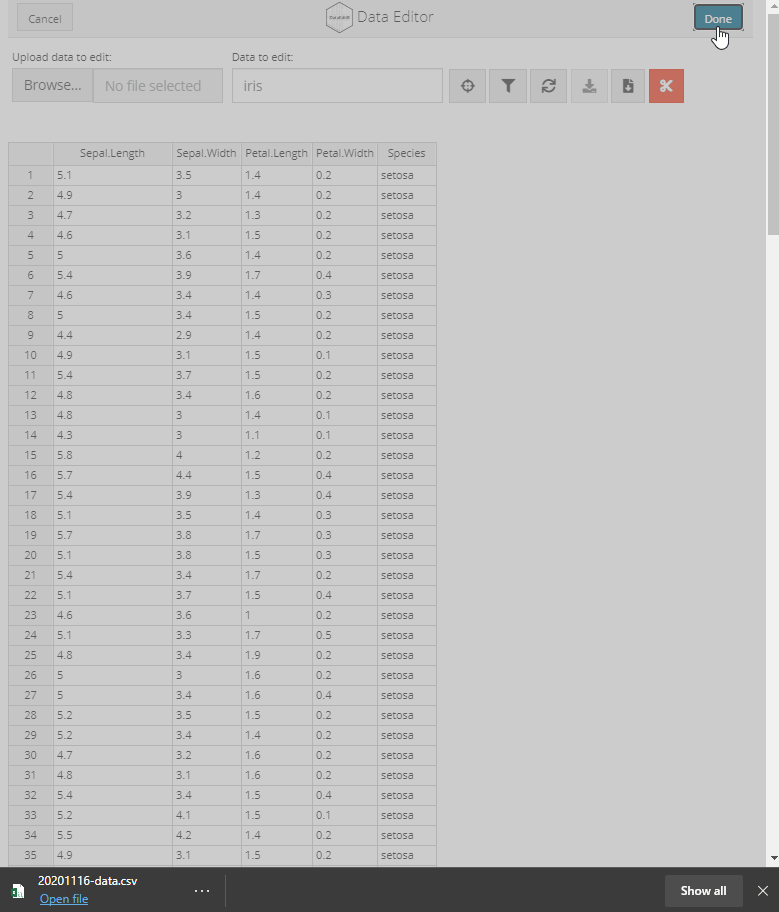
Manual data entry and editing in R can be tedious, especially if you have limited coding experience and are accustomed to using software with a Graphical User Interface (GUI). DataEditR is an R package built on shiny and rhandsontable that makes it easy to interactively view, enter, filter and edit data. If you are new to DataEditR visit https://dillonhammill.github.io/DataEditR/ to get started.
DataEditR can be installed from CRAN:
install.packages("DataEditR")The development version of DataEditR can be installed directly from GitHub:
library(devtools)
install_github("DillonHammill/DataEditR")To ensure that DataEditR works as expected, you will also need to
install my fork of rhandsontable:
devtools::install_github("DillonHammill/rhandsontable")DataEditR ships with a series of shiny modules, namely dataInput,
dataSelect, dataFilter, dataEdit and dataOutput which have been
wrapped up into a single function called data_edit() to create an
interactive data editor. You can use data_edit() as a standalone
application, or include the relevant modules within your own shiny
applications. Alternatively, DataEditR also ships with an RStudio
add-in should you prefer to interact with it in this way.
- RStudio add-in
- flexible display options (either
dialogbox,browseror RStudioviewerpane) - fast rendering to quickly view datasets
- ability to interactively create data.frames from scratch
- load tabular data saved to file using any reading function
(e.g.
read.csv()) - save edited data to file using any writing function
(e.g.
write.csv()) - return appropriately formatted data as an R object for downstream use
- code required to create edited data can be optionally printed to the console or saved to a file
- support for custom themes through
bslibpackage - customisable user interface (title, logo and modules)
- row indices are always displayed for easy navigation
- switch between datasets or files without having to leave the application
- column selection using the
dataSelectmodule - row selection using the
dataFiltermodule - edit row or column names
- addition or removal of rows or columns
- manual column resizing
- drag to fill cells
- copy or paste data to and from external software
- custom column types to simplify user input (e.g. checkboxes and dropdown menus)
- support for readonly columns to prevent users from editing certain columns
- control over which column names can be edited
- stretch columns horizontally to fill available space
- programmatically add columns or rows to data prior to loading into the data editor
A quick demonstration of some of these features can be seen below, where
we use data_edit() to make changes to the mtcars dataset and save
the result to a new csv file:
# Load required packages
library(DataEditR)
# Save output to R object & csv file
mtcars_new <- data_edit(mtcars,
save_as = "mtcars_new.csv")DataEditR is built using the fantastic rhandsontable package. DataEditR makes use of many features for entering and editing data, but rhandsontable has support for much more sophisticated interactive representations of data should you need them. The user interface of DataEditR has been inspired by the editData package which is a great alternative to DataEditR.
Please note that the DataEditR project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
If you use DataEditR in your work, please cite the package as follows:
citation("DataEditR")
#>
#> To cite package 'DataEditR' in publications use:
#>
#> Dillon Hammill (2022). DataEditR: An Interactive Editor for Viewing,
#> Entering, Filtering & Editing Data. R package version 0.1.5.
#> https://github.com/DillonHammill/DataEditR
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {DataEditR: An Interactive Editor for Viewing, Entering, Filtering & Editing Data},
#> author = {Dillon Hammill},
#> year = {2022},
#> note = {R package version 0.1.5},
#> url = {https://github.com/DillonHammill/DataEditR},
#> }