deltares / dfm_tools Goto Github PK
View Code? Open in Web Editor NEWA Python package for pre- and postprocessing D-FlowFM model input and output files
Home Page: https://deltares.github.io/dfm_tools/
License: GNU General Public License v3.0
A Python package for pre- and postprocessing D-FlowFM model input and output files
Home Page: https://deltares.github.io/dfm_tools/
License: GNU General Public License v3.0
For CMCC, HIRLAM, Delft3D4, WAQUA and other models, there are 2D lat/lon variables in the dataset (curvilinear grids, sometimes polar). Decide on interpolation method used in dfmt.interp_regularnc_to_plipoints(). ds.interp() does not work with 2D lat/lon vars (pydata/xarray#2281), so currently using KDTree for CMCC.
uds.ugrid.sel_points()Very maybe uds.ugrid.sel(x/y) could help out here (Deltares/xugrid#26). Or for instance implement uds.ugrid.isel(faces=idx_faces), where also edges/nodes are subsetted (Deltares/xugrid#32) >> fixed
If it would be possible subset an entire dataset (dims: faces, edges, nodes) based on faceidx, it would also be easier to remove ghostcells as in dfmt.open_partitioned_dataset() (#207)
xu.grid.intersect_edges: #252polyline_mapslice() now uses xu.ugrid2d.intersect_edges and sorts them. Put sorting in intersect_edges or numba.celltree (Deltares/xugrid#65 is probably related) >> this seems to be solved, but check more thoroughlycalcdist_fromlatlon is derived from hardcoded varnames wgs84 or projected_coordinate_system, maybe Deltares/xugrid#42 (but would still require conversion to cartesian/spherical). At least find coordvar based on attribute if possible.This should probably be aligned, all z-variables as coords or all as data_vars.
First check if this is also different in models that are run with new dflowfm kernel. If so, report as UNST issue.
Tasks:
Functions that currently raise a DeprecationWarning:
Functions that currently print a DeprecationWarning:
would consist of:
quantity and maybe analysistype from long_name (e.g. "temperature, average value"). Issue for manalysistype from varname (e.g. "mesh2d_fourier001_mean") instead (standard_name is not always present), this prevents unclarities with underscores/spaces. However, is there always an underscore present there?tstart/tstop datestrings from numstart+numstop+reftime. However, numstart/numstop are defined twice and would have to be merged: starttime_fourier_analysis_in_minutes_since_reference_date and starttime_min_max_analysis_in_minutes_since_reference_datef"{quantity}_{analysistype}_{tstart}_{tstop}" names would then probably make most sense. However, some quantities have spaces/underscores in them, so it might be better to have shorter names like ux/uy/wl etc.numcyc attr: xarray.Dataset.filter_by_attrsGetting Foreman tidal frequencies:
import pandas as pd
file_freqs = 'https://raw.githubusercontent.com/Deltares/hatyan/main/hatyan/data/data_foreman_frequencies.txt'
freqs = pd.read_csv(file_freqs,names=['freq','dependents'],delim_whitespace=True,comment='#')
print(freqs.loc['M2','freq'])
Used example files:
p:\archivedprojects\11203379-005-mwra-updated-bem\03_model\02_final\A72_ntsu0_kzlb2\DFM_OUTPUT_MB_02_fou\MB_02_0000_fou.ncp:\1230882-emodnet_hrsm\GTSMv3.0EMODnet\EMOD_MichaelTUM_yearcomponents\GTSMv4.1_yeartide_2014_2.20.06\output\gtsm_model_0000_fou.ncAdd kml export support, for instance like:
Improve dfmt.open_partitioned_dataset():
This might fix the issue of reprojecting a uds as in the header of https://github.com/Deltares/dfm_tools/blob/main/tests/examples_workinprogress/workinprogress_grid_convert_coordinates.py
This should fix improper edges plotting in https://github.com/Deltares/dfm_tools/blob/main/tests/examples_workinprogress/workinprogress_plot_edges.py
This might fix #203
Opening large mapfiles takes quite some time, this might for instance be because of the decoding of time etc. This could also be done only once, after merging of the mapfile. However, the stuff that takes time is cached so the second opening is more than 10 times faster. Beware the performance for the second opening does not get less.
Some timings:
- DCSM 3D 20 partitions 367 timesteps: 219.0 sec
- RMM 2D 8 partitions 421 timesteps: 60.6 sec
- GTSM 2D 8 partitions 746 timesteps: 73.8 sec
- RMM 3D 40 partitions 146 timesteps: 166.0 sec
- MWRA 3D 20 partitions 2551 timesteps: 826.2 sec
Next release (4.2.0 or 5.0.0):
Test GTSM-specific new features:
mesh2d_refine_based_on_gridded_samples() API now supports multiple dtypes (Deltares/MeshKernelPy#146). Check if gtsm refinement with meshkernelpy is possible with coarsefac 4 >> short/float distinction >> test gtsm memory consumtion for two dtypes (with release)Next:
Other:
dfm_tools/dfm_tools/meshkernel_helpers.py
Line 19 in 14fc1ae
Related issues:
connect_cells separately (without refining)?p:\dflowfm\maintenance\JIRA\06000-06999\06548\meshkernel_interp.py, can be used to interpolate bathy to?Workflows:
DCSM steps:
mesh2d_delete_small_flow_edges_and_small_triangles (#812 (comment))mesh2d_delete_hanging_edgesmesh2d_merge_nodesRelease 3.0.0:
xugrid.ugrid.ugrid1d.Ugrid1d.meshkernel and xugrid.ugrid.ugrid2d.Ugrid2d.meshkernel both call mk.MeshKernel(is_geographic=False) so that now failsmk 4.1.0 (released 2024-02-15):
Shapely minimal version is tested (shapely>=1.7.0), put this in requirements instead
When hydrolib-core 0.5.0 and meshkernel 2.0.3 is available, update deps to:
TODO in example scriptsTest with new env from yml.
Recompute like suggested in ArcticSnow/TopoPyScale#60 (comment)
Implementation: https://github.com/ArcticSnow/TopoPyScale/blob/494f4e7ea17830ba3d23627bf22ee200a6c4f082/TopoPyScale/topo_export.py#L21
Currently installing an older numpy version to avoid "SystemError: initialization of _internal failed without raising an exception"
This happens in the generate-documentation actions 57 to 61 and in binder, but in general upon creation of dfm_tools_env from environment.yml.
This post suggests to restrict the numpy version and using "numpy<1.24" indeed resolves the issue.
https://stackoverflow.com/questions/74947992/how-to-remove-the-error-systemerror-initialization-of-internal-failed-without
Release this restriction when the dependency conflict is resolved by numba: numba/numba#8464
Also:
classifiers and requires-python)xarray.to_netcdf() of opened mfdataset results in incorrect data when not using manual encoding
import os
import xarray as xr
import matplotlib.pyplot as plt
plt.close('all')
#open data
dir_data = r'p:\11207892-pez-metoceanmc\3D-DCSM-FM\workflow_manual\01_scripts\04_meteo\era5_temp'
file_nc = os.path.join(dir_data,'era5_mslp_*.nc')
data_xr = xr.open_mfdataset(file_nc)
#optional encoding
#data_xr.msl.encoding['dtype'] = 'float32' #TODO: updating dtype in encoding solves the issue. Source data is int, opened data is float, but encoding is still int.
#data_xr.msl.encoding['_FillValue'] = float(data_xr.msl.encoding['_FillValue'])
#data_xr.msl.encoding['missing_value'] = float(data_xr.msl.encoding['missing_value'])
#data_xr.msl.encoding['zlib'] = True #no effect
#data_xr.msl.encoding['scale_factor'] = 0.01
#data_xr.msl.encoding['add_offset'] = 0
#write to netcdf file
file_out = os.path.join('era5_mslp_out.nc')
data_xr.to_netcdf(file_out)
fig,(ax1,ax2) = plt.subplots(1,2,figsize=(11,5))
data_xr.msl.sel(time='2023-01-24 02:00:00').plot(ax=ax1,cmap='jet') #original dataset
with xr.open_dataset(file_out) as data_xr_check:
data_xr_check.msl.sel(time='2023-01-24 02:00:00').plot(ax=ax2,cmap='jet') #written dataset
fig.tight_layout()
This results in incorrect data in the written file (right):
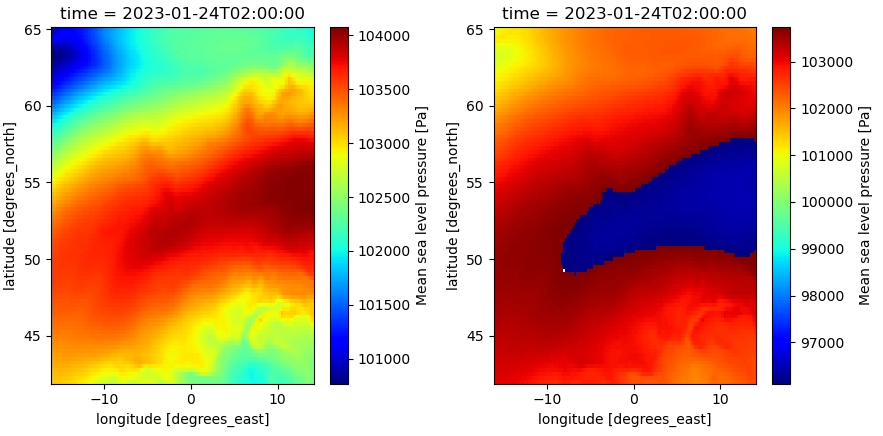
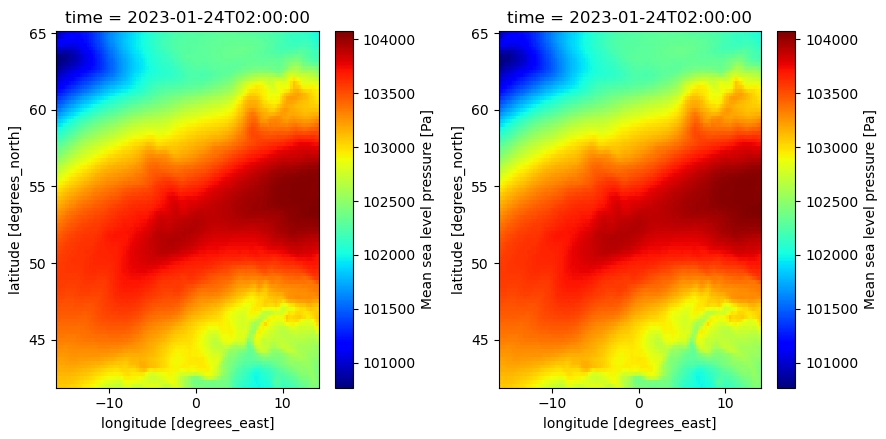
When updating the dtype (from int to float) in the variable encoding, this issues is solved:
The encoding in the source dataset:
data_xr.msl.encoding
Out[28]:
{'source': 'p:\\11207892-pez-metoceanmc\\3D-DCSM-FM\\workflow_manual\\01_scripts\\04_meteo\\era5_temp\\era5_mslp_2022-11.nc',
'original_shape': (720, 93, 121),
'dtype': dtype('int16'),
'missing_value': -32767,
'_FillValue': -32767,
'scale_factor': 0.11615998809759968,
'add_offset': 99924.34817000595}
Possible issue: source data is integers, but opening files with different scaling_factors (from different files) converts it to floats (or maybe this always happens). The dtype in the encoding is still int, so this is how the netcdf is written, but probably something does not fit within the int-bounds.
First setup: https://github.com/Deltares/dfm_tools/blob/main/tests/examples_workinprogress/workinprogress_nestingFMtoFM.py
Also request from RS/PK to add Neuman boundary, where inflowing direction of grid should be known (possible?). Or alternatively, combination of waterlevel/ux/uy boundary, which is simpler to implement.
Related, nesting SFINCS in FM is done in GC with:
Also request from WO to add Riemann boundary, so combination of waterlevel and discharge (latter from cross-sections). For morphology we also require nesting bed level changes (form obsfiles) and sediment transport (form cross-sections).
Also align with nesting in ocean models like CMEMS, maybe also consider nesting in other existing models (shyfem, schism, delft3d4, etc).
Removing deprecated code makes https://github.com/Deltares/dfm_tools/blob/main/tests/examples/preprocess_ini_rst_nc_to_xyz.py fail, since it reads rst files which are in the unsupported mapformat=1. Add preprocess function to generate mesh2d variable with pointers to relevant topology variable.
The internal structure of datasets opened with dfmt.open_partitioned_dataset() is not consistent. This might be because edges/nodes are not ghostcell-filtered or reindexed.
import xugrid as xu
import dfm_tools as dfmt
file_nc = r'c:\DATA\dfm_tools_testdata\DFM_3D_z_Grevelingen\computations\run01\DFM_OUTPUT_Grevelingen-FM\Grevelingen-FM_0000_map.nc'
data_frommap = xu.open_dataset(file_nc)
data_frommap.ugrid.to_netcdf('test.nc') #this works
data_frommap2 = dfmt.open_partitioned_dataset('test.nc') #this also works
data_frommap = dfmt.open_partitioned_dataset(file_nc)
data_frommap.ugrid.to_netcdf('test.nc') #TODO: "ValueError: cannot reindex or align along dimension 'mesh2d_nEdges' because of conflicting dimension sizes: {77761, 67906}"
#data_frommap2 = dfmt.open_partitioned_dataset('test.nc') #not tested yet
Tasks for first implementation:
status = os.system('dir')For FM, there are several file types for bathymetry and serveral packages that could read/write them:
import gdal or from osgeo import gdal (does not work in dfm_tools_env for some reason)Checks for next release (may 2024):
From hydromt_delft3dfm:
Other:
/ is not read but also raises no error: Deltares/HYDROLIB-core#485 (comment) (same for missing keywords)Can be closed? (dfm_tools/xugrid alternatives):
ds.sel(station='stationname') ValueError: multi-index does not support 'method' and 'tolerance'. Alternative method is via KDTreeds.sel(x=slice(),y=slice() >> TypeError: float() argument must be a string or a number, not 'slice'. select with polygon would also result in bool, so might not matterhydrolib-core issues, to be included in 0.5.0:
TimModel_to_Dataset when donedfmt.scatter_to_regulargrid() was replaced by dfmt.rasterize_ugrid(), which is a working implementation for a new regridder based on the 500x500 regridder in xugrid for uda.ugrid.plot.imshow() (Deltares/xugrid#31). The rasterization is used in e.g. workinprogress_mapfile_to_regulargrid.py and postprocess_mapnc_ugrid.py
Left tasks:
moved to #215
Deprecate dfmt.get_ncmodeldata() and related functions, these have "DeprecationWarning" in these scripts:
Currently results in an error: AttributeError: 'Dataset' object has no attribute 'set_crs'
It was added to xr.UgridDataset via the ugrid accessor. Consider doing something similar for hisfiles.
Currently added a xugrid uds.grid.plot() method via the init.py file of dfm_tools. This is not necessary anymore, since Deltares/xugrid#28 and Deltares/xugrid#54 are closed.
Install toolbox from conda .yml file
did: conda env update -f newenvironment.yml
with following lines in yml that give error
dependencies:
- pip:
- git+https://github.com/openearth/dfm_tools.git@6ac91e323ad9228cd10903a3e6af4ac15ad72b20
error:
File "\dfm_tools\setup.py", line 8, in <module>
readme = readme_file.read()
File "\lib\codecs.py", line 322, in decode
(result, consumed) = self._buffer_decode(data, self.errors, final)
UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 3617: invalid start byte
similar error will be found later in the history file
seamless installation ;)
This is just a test issue to see if it works.
First branch has been created and there is a howto_git.txt explaining the theoretical way to work with it. Please check if it works
Input quantity for Boundary should be the bcvarname as is correctly specified in get_conversion_dict in interpolate_grid2bnd.py. E.g. salinity should become salinitybnd.
boundary_object = Boundary(quantity=quantity, #TODO: nodeId / bndWidth1D / bndBlDepth are written as empty values, but they should not be written if not supplied. https://github.com/Deltares/HYDROLIB-core/issues/319
locationfile=Path(dir_out,file_pli.name),
forcingfile=ForcingModel_object,
)
Could land in xugrid, dfm_tools, meshkernel or hydrolib-core.
Actions like >> current package:
Temporary fix for cells that go "around the back" of global models. Later properly fix in xugrid: Deltares/xugrid#63
uds.sel() to drop all other faces. Possible with multiple polygons?0.8.1:
Soon:
Then:
Later:
xugrid 0.7.0 or older:
"xugrid\ugrid\ugridbase.py:326: RuntimeWarning: invalid value encountered in cast cast = data.astype(dtype, copy=True)" >> Deltares/xugrid#101SKIP_test_xugrid_opendataset_ugridplot_contourf() and other testcase since it is tested in xugrid testbank now. Also uncomment contour/contourf in ipynb and ugrid example script.xr.concat in xarray 2023.2.0 to 2023.5.0:Deltares/xugrid#106Todo:
test_ssh_catalog_subset())from observations import *Follow-up: #712
variables = [var for var in ds.data_vars if set(["layer", "time", grid.face_dimension]).issubset(ds[var].dims)]Fix failing testbank cases (24-01-2023).
_____________________________ test_run_examples[preprocess_meteo_mergenetCDFtime_xarray] ______________________________
C:\DATA\dfm_tools\tests\test_dfm_tools.py:43: in test_run_examples
------------------------------------------------ Captured stdout call -------------------------------------------------
opening multifile dataset of 180 files matching "era5_.*(chnk|mslp|u10n|v10n)_.*\.nc" (can take a while with lots of files)
------------------------------------------------ Captured stderr call -------------------------------------------------
Traceback (most recent call last):
[...]
OSError: [Errno -51] NetCDF: Unknown file format: b'p:\\metocean-data\\open\\ERA5\\data\\Irish_North_Baltic_Sea\\u10n\\era5_u10n_1992.nc'
______________________________________ test_zlayermodel_correct_layers_THISFAILS ______________________________________
C:\DATA\dfm_tools\tests\test_dfm_tools.py:165: in test_zlayermodel_correct_layers_THISFAILS
assert (np.abs(vals_zcc_top-vals_wl)<1e-6).all() #this should pass
E AssertionError: assert False
E + where False = <built-in method all of numpy.ndarray object at 0x00000220BFA8E570>()
E + where <built-in method all of numpy.ndarray object at 0x00000220BFA8E570> = array([2.5124 , 2.2804 , 2.2921 , ..., 2.284 , 2.25245, 2.25245]) < 1e-06.all
E + where array([2.5124 , 2.2804 , 2.2921 , ..., 2.284 , 2.25245, 2.25245]) = <ufunc 'absolute'>((array([-0.875, -0.875, -0.875, ..., -0.875, -0.875, -0.875]) - array([1.6374 , 1.4054 , 1.4171 , ..., 1.409 , 1.37745, 1.37745])))
E + where <ufunc 'absolute'> = np.abs
Deprecate dfmt.get_ugrid_verts(). Since Deltares/xugrid#48 was solved, this can be replaced with uds.grid.face_node_coordinates everywhere. Maybe first add Deprecationwarning, and/or put new code in function for now.
Convert FM data (map/grid/obspoints/thd/pli/etc) to kml/shp, including coordinate conversion
Shapefile example (kml was also added for grid): https://github.com/Deltares/dfm_tools/blob/main/tests/examples_workinprogress/workinprogress_exporttoshapefile.py
KML example for obspoints: #808
Also convert obs/crs/etc to shape/kml
Maybe geowomat: https://geowombat.readthedocs.io/en/latest/api/geowombat.core.geoxarray.GeoWombatAccessor.html
The plot_ztdata function uses pcolormesh to plot the data value against a 2D mesh of time and depth. The time and depth arrays are masked, which is not supported by pcolormesh. Using pcolor instead seems to get around the issue.
I suggest replacing line #837 of get_nc.py with:
pc = ax.pcolor(time_mesh_cor, data_fromhis_zcor_flat, dfmtools_hisvar_flat, **kwargs)
Documentation/license:
Code style/quality:
Create issues:
Testbank:
requiresdata (reduces codecov but that is ok)requireslocaldata tests by making testdata available (and avoiding p-drive links). Now we have opendap download. Maybe use HYDROLIB-data repos instead (or xugrid cached downloads)? Maybe use dsctestbench data on repos, how to authenticate on github? At least make cds/cmems testcases work on github by setting env varsA declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.