The anomalize package functionality has been superceded by timetk.
We suggest you begin to use the timetk::anomalize() to benefit from
enhanced functionality to get improvements going forward. Learn more
about Anomaly Detection with timetk
here.
The original anomalize package functionality will be maintained for
previous code bases that use the legacy functionality.
To prevent the new timetk functionality from conflicting with old
anomalize code, use these lines:
library(anomalize)
anomalize <- anomalize::anomalize
plot_anomalies <- anomalize::plot_anomaliesTidy anomaly detection
anomalize enables a tidy workflow for detecting anomalies in data. The
main functions are time_decompose(), anomalize(), and
time_recompose(). When combined, it’s quite simple to decompose time
series, detect anomalies, and create bands separating the “normal” data
from the anomalous data.
Check out our entire Software Intro Series on YouTube!
You can install the development version with devtools or the most
recent CRAN version with install.packages():
# devtools::install_github("business-science/anomalize")
install.packages("anomalize")anomalize has three main functions:
time_decompose(): Separates the time series into seasonal, trend, and remainder componentsanomalize(): Applies anomaly detection methods to the remainder component.time_recompose(): Calculates limits that separate the “normal” data from the anomalies!
Load the anomalize package. Usually, you will also load the tidyverse
as well!
library(anomalize)
library(tidyverse)
# NOTE: timetk now has anomaly detection built in, which
# will get the new functionality going forward.
# Use this script to prevent overwriting legacy anomalize:
anomalize <- anomalize::anomalize
plot_anomalies <- anomalize::plot_anomaliesNext, let’s get some data. anomalize ships with a data set called
tidyverse_cran_downloads that contains the daily CRAN download counts
for 15 “tidy” packages from 2017-01-01 to 2018-03-01.
Suppose we want to determine which daily download “counts” are
anomalous. It’s as easy as using the three main functions
(time_decompose(), anomalize(), and time_recompose()) along with a
visualization function, plot_anomalies().
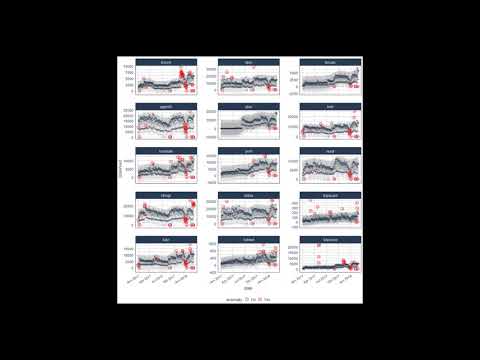
tidyverse_cran_downloads %>%
# Data Manipulation / Anomaly Detection
time_decompose(count, method = "stl") %>%
anomalize(remainder, method = "iqr") %>%
time_recompose() %>%
# Anomaly Visualization
plot_anomalies(time_recomposed = TRUE, ncol = 3, alpha_dots = 0.25) +
ggplot2::labs(title = "Tidyverse Anomalies", subtitle = "STL + IQR Methods") Check out the anomalize Quick Start
Guide.
Yes! Anomalize has a new function, clean_anomalies(), that can be used
to repair time series prior to forecasting. We have a brand new
vignette - Reduce Forecast Error (by 32%) with Cleaned
Anomalies.
tidyverse_cran_downloads %>%
dplyr::filter(package == "lubridate") %>%
dplyr::ungroup() %>%
time_decompose(count) %>%
anomalize(remainder) %>%
# New function that cleans & repairs anomalies!
clean_anomalies() %>%
dplyr::select(date, anomaly, observed, observed_cleaned) %>%
dplyr::filter(anomaly == "Yes")
#> # A time tibble: 19 × 4
#> # Index: date
#> date anomaly observed observed_cleaned
#> <date> <chr> <dbl> <dbl>
#> 1 2017-01-12 Yes -1.14e-13 3522.
#> 2 2017-04-19 Yes 8.55e+ 3 5202.
#> 3 2017-09-01 Yes 3.98e-13 4137.
#> 4 2017-09-07 Yes 9.49e+ 3 4871.
#> 5 2017-10-30 Yes 1.20e+ 4 6413.
#> 6 2017-11-13 Yes 1.03e+ 4 6641.
#> 7 2017-11-14 Yes 1.15e+ 4 7250.
#> 8 2017-12-04 Yes 1.03e+ 4 6519.
#> 9 2017-12-05 Yes 1.06e+ 4 7099.
#> 10 2017-12-27 Yes 3.69e+ 3 7073.
#> 11 2018-01-01 Yes 1.87e+ 3 6418.
#> 12 2018-01-05 Yes -5.68e-14 6293.
#> 13 2018-01-13 Yes 7.64e+ 3 4141.
#> 14 2018-02-07 Yes 1.19e+ 4 8539.
#> 15 2018-02-08 Yes 1.17e+ 4 8237.
#> 16 2018-02-09 Yes -5.68e-14 7780.
#> 17 2018-02-10 Yes 0 5478.
#> 18 2018-02-23 Yes -5.68e-14 8519.
#> 19 2018-02-24 Yes 0 6218.There are a several extra capabilities:
plot_anomaly_decomposition()for visualizing the inner workings of how algorithm detects anomalies in the “remainder”.
tidyverse_cran_downloads %>%
dplyr::filter(package == "lubridate") %>%
dplyr::ungroup() %>%
time_decompose(count) %>%
anomalize(remainder) %>%
plot_anomaly_decomposition() +
ggplot2::labs(title = "Decomposition of Anomalized Lubridate Downloads")For more information on the anomalize methods and the inner workings,
please see “Anomalize Methods”
Vignette.
Several other packages were instrumental in developing anomaly detection
methods used in anomalize:
- Twitter’s
AnomalyDetection, which implements decomposition using median spans and the Generalized Extreme Studentized Deviation (GESD) test for anomalies. forecast::tsoutliers()function, which implements the IQR method.
Business Science offers two 1-hour courses on Anomaly Detection:
-
Learning Lab 18 - Time Series Anomaly Detection with
anomalize -
Learning Lab 17 - Anomaly Detection with
H2OMachine Learning