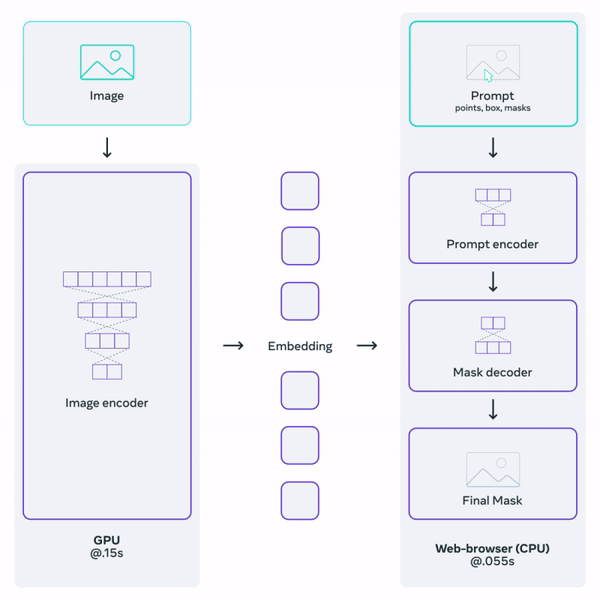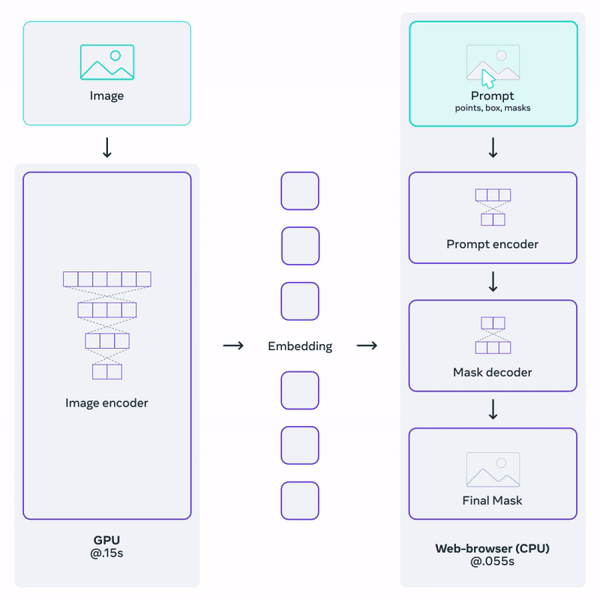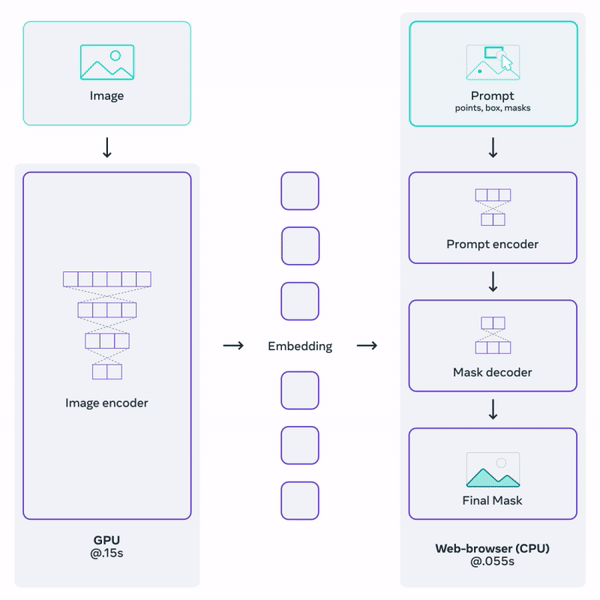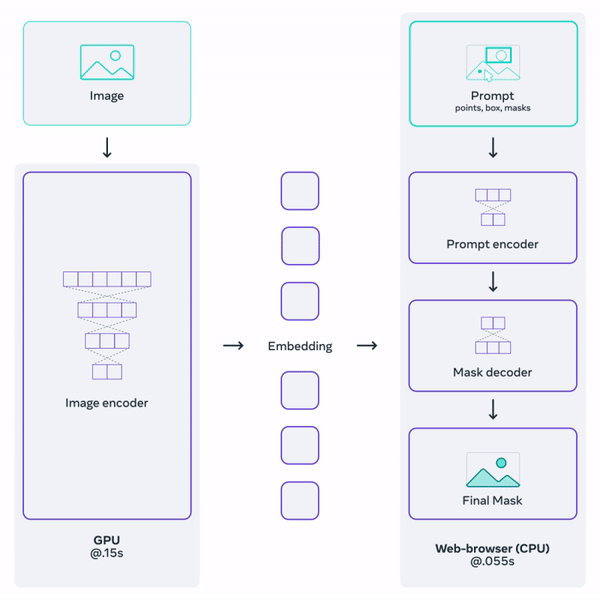
Histology (microscopy) data is widely used by neuropathologists to study demylienation in the nervous system. This project aims to leverage a general-purpose foundation model to segment myelin on histology images. Foundation models are large DL models trained on large-scale data. They learn a general representation that can be adapted to a variety of downstream tasks. OpenAI's GPT serie, for example, are examples of foundation models for natural language processing. Facebook's Segment-Anything-Model (SAM) is one such promptable foundation model for segmentation tasks.
The data used for this project is the data_axondeepseg_tem dataset privately hosted on an internal server with git-annex. It was used to train this model. It's also our biggest annotated dataset for myelin segmentation (20 subjects, 1360 MPx of manually segmented images). An older version of this dataset is publicly available on this OSF repository, under the data/raw/ directory. For more information on how to acces the data, see the How to reproduce section below.
[...]
For a complete guide to reproduce these results, please see the README in the scripts folder.